(9-13)Summary of Group Meeting (09.13.2013)
v Discussions on manuscript_PLOS Computational BiologyAccording to Dr Raj’s suggestions, Ping’s comments, and new published literatures, our manuscript need to be future edited as following:
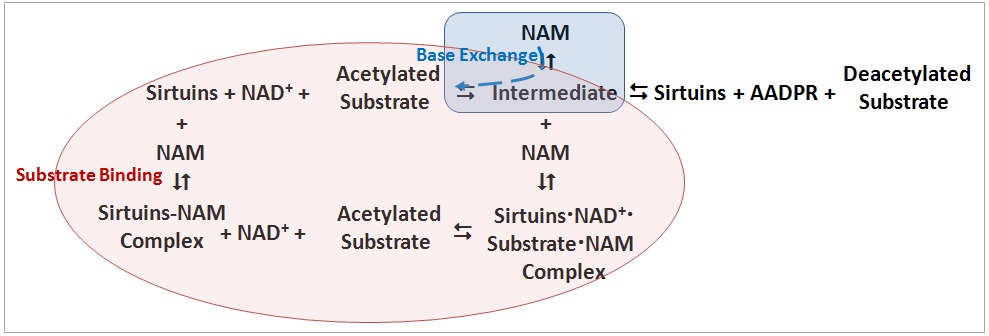
1. Two NAM inhibition mechanisms were observed (as shown in figure below): (A) during substrate (NAD+) binding and (B) through base exchange reaction.
In our manuscript, we want to address is the following issues:
(a) our computational studies focus on the inhibition mechanism A for both Sir2 and SIRT3 by evaluating the binding modes of NAD+;
(b) Mechanisms A and B may co-exist, and NAM plays different roles.
(c) Mechanism B is specific to NAM, and isoNAM activation is achieved by NAM inhibition relief in this mechanism.
2. Analysis of competitive & noncompetitive inhibition modes
(a) A concept of “mixed noncompetitive inhibition” was introduced.
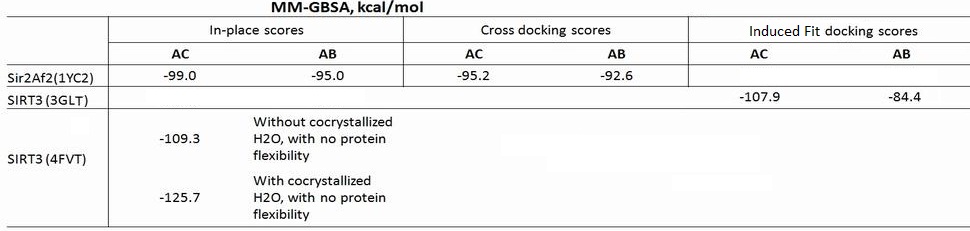
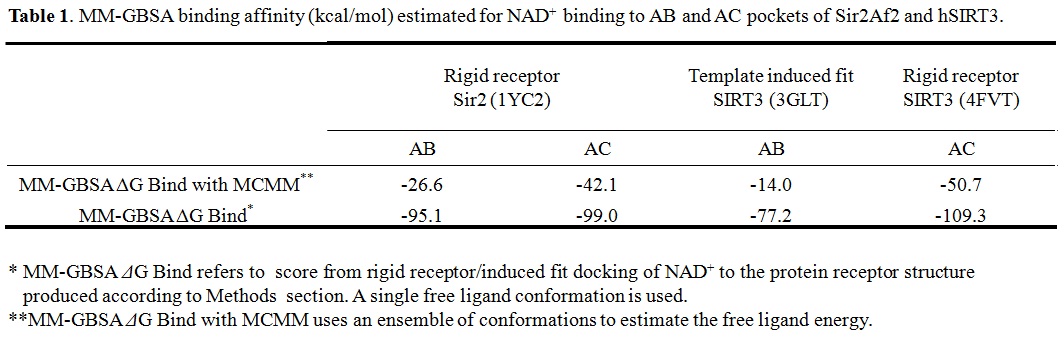
(b) For Sir2: our simulation results shows the MM-GBSA/MM-GBSA with MCMM scores of NAD+ for AB and AC pocket are both significant (-95.1 and -99.0 kcal/mol in MM-GBSA, and -26.6 and -42.1 kcal/mol in MM-GBSA with MCMM), which indicated NAM binding did not affect NAD+ binding much. In this case, the mixed noncompetitive inhibition is close to pure noncompetitive inhibition.
(c) For SIRT3: MM-GBSAAC>> MM-GBSAAB (refer to -109.3 and -77.2 kcal/mol, and -50.7 Vs. -14.0 kcal/mol in MM-GBSA with MCMM) indicated that the binding of NAD+ to AB pocket is unfavorable compared to other binding modes. The binding of NAM in AC pocket significantly decreased the ability of NAD+ binding to AB pocked. In this case, the mixed noncompetitive inhibition acts close to competitive inhibition.
(d) New table will be included.
3. PNAS paper_Gertz et al. Ex-527 inhibits sirtuins by exploiting theunique NAD+-dependent deacetylation mechanism. PNAS (2013) E2772-E2781.
(a) Need to structurally confirm if and how
i. Ex-527 interacts with NAD+ in Sir2 and SIRT3
ii. Ex-527 interacts with intermediate in Sir2 and SIRT3
(b) In the manuscript, we need to address that Ex-527 noncompetitively inhibits SIRT3 by a different inhibition mechanism. Ex-527 occupies the NAM site and contacts the ribose of NAD+ or the coproduct 1O-ADDP ribose. The direct contact of Ex-527 and intermediate stabilized the Ex-527-substrate-enzyme complex, therefore inhibits the enzyme conformational change for releasing product.
(c) Take home lesson is interaction with the intermediate, which is undesirable for activator design. Thus if we are screening molecules that might eventually be used as leads for activators, we want to screen out the type of binding that was similar toEx-527.
4. XG_provide a time frame for experimental work in the lab in terms of
(a) Investigate inhibition mode of existing potent inhibitors, such as salermide (IC50_SIRT3=27uM), AC93253 (IC50_SIRT3=18 uM). May repeat Ex-527 to see if our results agree with reported data. How long will take, how much money need to spend for purchasing Fluor-de-Lys kit
(b) Continuous assay, how far need to go, if we have it how long for each assay and how much money will spend
XG(9-20):
Schedule_ongoing experiments. (gray is done.)
(1) Continuous enzyme coupled assay.
- PCR amplification of PNCA from Samonella genomic DNA (Primer design, PCR condition optimization, …)
- Subcloning into appropriate expression vector-pGEX6P3. (TOPO cloning, transformation of TOP10 chemical competent cell, identification of positive clones, Mini and Midiprep for sequencing, Digestion, Alkaline phosphatase, Calf Intestinal_CIP for Ligation, …)
- Plasmid maxi preparation
- Linearize the constructs
- Transformation the linear construct into the appropriate host strain BL21(DE3)
- Confirmation of positive transformants
- Expression optimization and confirmation. (Screening of transformants, media formulation and inducer concentrations, induction temperature and length, culture lysate conditions, SDS-PAGE gel, …)
- Large-scale expression. (1 week)
- Purification (two weeks) and confirmation (western blot/HPLC: 1 week).
- Activity measurements (1-2 weeks)
Week 1: Inhibitor physiochemical properties
(a) Solubility in H2O, assay buffer
(b) If not soluble or have very poor solubility in assay buffer, selection of organic solvent
(c) Concentration range, pH range
(d) If has color, background minimization
Week 2: Standardization of working assay systems (Fluor-de-Lys and Continuous Enzyme Coupled Assay)
- Standard calibration curve
- Titration of developer signal stabilization
- Measurement of Km(NAD+) and Km(substrate peptide)
- IC50 (may try 10 different [inhibitor] then narrow down for specific range.)
- Ki ( [NAD+]=0, 62.5, 125, 250, 500, 1000, 1500, 3000 uM with different [inhibitor].)
In summary
For inhibitor screening (IC50 measurements to show the potency of inhibition, Ki then can be calculated), it takes 1 month for testing 5 inhibitors using Fluor-de-Lys SIRT3 drug discovery kit (2 kits, ~$1200), but 10 inhibitors using continuous enzyme coupled assay (Costs of the chemical reagents used in assay, like buffer solutions, chemicals, 96-well plates et al).
For inhibition mode study (determination of competitive, noncompetitive, uncompetitive inhibition modes), it takes 4 weeks for testing 1 inhibitor using Fluor-de Lys SIRT3 drug discovery kit (3 kits, ~$1800), but 3 weeks using continuous enzyme coupled assay (Costs of the chemical reagents used in assay, like buffer solutions, chemicals, 96-well plates et al).
5. XG_look for the possibility of the outsources for crystallography
XG(9-20): Two companies and a university core facility provide such a service. The details are listed below
GenScript USA Inc.
860 Centennial Ave
Piscataway, NJ 08854
Email:[email protected]
CrystalPro Gnen –to-Structure Services
Phase I: High purity protein production
Phase II: Crystallization and structure determination
Phase III: Protein-compound complex structure determination
SHAMROCK STRUCTURES, LLC
1440 Davey Road
Woodridge, IL 60517
Email: [email protected]
Shamrock Structures provides protein crystallography services on an outsourced basis to pharmaceutical companies. Our clients engage with us to apply technology and expertise in X-ray crystallography to solve protein structures. Most often, our clients seek co-crystal structure determination, meaning their lead drug compound(s) in complex with a protein target of interest.
Shamrock Structures are advantageously located a mile from the U.S. Department of Energy’s Argonne National Laboratory and we have an agreement with Argonne to efficiently access the Advanced Photon Source (APS). The APS technology is the only 3rd generation X-ray synchrotron light source in North America and it permits the fastest protein structure determination currently possible.
X-ray Crystallographic Laboratory
Department of Chemistry
College of Science & Engineering
University of Minnesota
192C Kolthoff Hall,
207 Pleasant Street S. E.Minneapolis, MN 55455 E-mail: [email protected]
The XCL accepts samples for structural analysis from colleges and universities, as well as from industry. Clients of our facility receive by email a full report file and a CIF (crystallographic information file) which is required by most journals.
6.Ping_ Analyze the inhibitor library Xiangying has and pick the most representative molecules for experiments.
(8-12) The related references have been uploaded in Dropbox/PMC-AT Research/References.
References for "calculation of protein-ligand binding affinities" (8-9)
Flexibility and binding affinity in protein_Tuffery_2012.pdfBinding free energy calculations and biologicaltesting of novel thiobarbiturates as inhibitors_Uciechowska_2012.pdf
Are scoring functions in protein-protein docking ready to predict interactomes_clues from a novel binding affinity benchmark.pdf
XG(7-3): The JMB cover letter is uploaded. Please feel free to modify it. JMB cover letter.docx
EK(6-27): Excel spreadsheet with the updated cross-docking for Sir2. See columns G and H.
EK(6-19): Answers to the following from the task list are provided
- (7) Insert and discuss Eric’s new figure showing comparison of AB pockets for Sir2/SIRT3. If he highlights specific interactions, mention them. Mention RMSD, but do not mention scores unless we can show these individual interaction energies are similar in magnitude in Sir2/SIRT3 AB.
- see EK(6-18) below.
- RMSD between NAD+ in Sir2 and SIRT3
- 2.1880 Å RMSD 'in place' where no ligand atoms are moved to optimize RMSD. Only the backbone protein atoms are superimposed, then an 'in place' RMSD of the ligand atoms are taken.
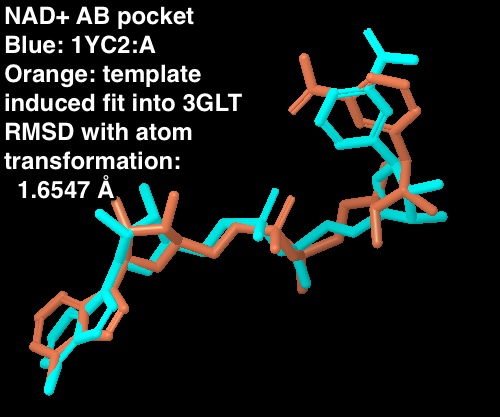
- 1.6547 Å RMSD when NAD+ atoms are transformed to optimize the RMSD. This is a measure of how similar the two NAD+ poses are irrespective of the protein atoms.
- Details about structures: the Sir2 NAD+ is from the 1YC2 chain A cocrystallized structure. The SIRT3 NAD+ is from the template induced fit docked into the AB pocket of SIRT3 (3GLT; entry 605 from maestro project redock.after.prime.prj where protein atoms were kept frozen in the final MM-GBSA rescoring)
- Note: there is also another co-crystallized structure of Sir2 with NAD+ in the AB pocket (1YC2 chain D, as apposed to chain A above). This co-crystallized structure has NAD+ in a slightly closer conformation to the docked SIRT3 NAD+. The main structural difference is the direction of the amide at the nicotinamide end; it is pointed inward in 1YC2:D (like the docked SIRT3 structure), but outward in 1YC2:A. 'In place' RMSD is 1.7946 Å, and 1.1428 Å when NAD+ atoms are transformed to optimize the RMSD. 1YC2:A with the slightly higher RMSD is reported above because this structure has a better score for MM-GBSA.
- (8) CHECK IF THE FOLLOWING ARE HIGHLIGHTED IN ERIC’S NEW FIG (task 5)
"Both Sir2 and hSIRT3 make similarly energetically favorable interactions in the AB pose, as well as in the AC [EK2] pose. The adenine and diphosphates have similar intermolecular interactions in the A pocket, especially with conserved residues. For example, conserved residues SER193 and SER321 form critical contacts with a phosphotidyl oxygen in Sir2 and SIRT3, respectively. As with the NAM in the C pocket cocrystallized structure of Sir2 and the docked structure in SIRT3, the carboxamide at the nicotinamide end of NAD+ in the AC binding mode makes a crucial hydrogen bond with Ile102 and Ile230 in Sir2 and SIRT3, respectively."- The first sentence above refers to the AB pose in Sir2 (cocrystallized 1YC2:A) and the docked SIRT3 (template induce fit into 3GLT). SER193 and SER321 appear in the same location in both structures, and both make at least one H-bond with a phosphotidyl oxygen. However, the two SER contacts are slightly different in the two structures. See this comment below. You could also mention other residues which are in similar locations and make similar contacts when comparing the two structures. See this table and the figures which show those similarities below.
- The second sentence that begins with 'As with the NAM...' refers to the AC binding mode. There is no figure in the paper nor the new task5 figures which show this for the C pocket.
- (10) Desirable to show a sequence alignment of Sir2/SIRT3 around AB pocket highlighting the homologous interactions and also the residues responsible for differences in backbone structure.
- Multiple Sequence Alignment of B-pocket residues that template induced fit moved.
 ppt of same (so that have text as well as jpg)multiple_sequence_alignment_showing_B-pocket_residues_that_template_induced_fit_moved.ppt
ppt of same (so that have text as well as jpg)multiple_sequence_alignment_showing_B-pocket_residues_that_template_induced_fit_moved.ppt
- Above figure: Alignment of B-pocket residues that template induced fit moved (SIRT3 Residues 320-324). Residues that moved in SIRT3 are highlighted in red. Many are highly conserved. The sequence alignment is from the Clustal 2.1 multiple sequence alignment of Human SIRT1 – SIRT7 and Sir2Af2, Sir2Tm. Algorithm: ClustalW2 http://www.ebi.ac.uk/Tools/msa/clustalw2/. Conserved residues indicated in the last row of the table: '*'== identical for all sequences ':'== very similar '.'== similar
- Above does not show ALL residues that are in the B-pocket. It only shows the ones in the sequence near those that moved (320-324). Below are additional figures showing other residues not near the 320-324 sequence numbers that are also in the B-pocket.


- The following multiple sequence alignment of SIRT1-7 and Sir2 highlight residues in the B-pocket of Sir2 and SIRT3.
- Multiple Sequence Alignment with SIRT3 B-pocket residues.doc
- XG (06-21):Since for JMB, there is a 8-Figure limitation for research article. I combined the figures based on task #5. I am not sure the following figure shows too much info.Figure caption: Figure X: Intermolecular protein-ligand interaction diagrams of (a) NAD+ cocrystallized in the AB pocket of Sir2 (1YC2 chain A). (b) NAD+ template induced fit into the AB pocket of SIRT3 (3GLT). In this flattened 2D representation, residues within 2.8 Å of the NAD+ are represented as colored spheres, where: red=acidic, green=hydrophobic, blue=polar, light gray=(Gly). Solid pink lines are H-bonds to the protein backbone; dotted pink are H-bonds to the side chains. Solvent exposed ligand atoms are shaded gray. Residues with 5 Å of the nicotinamide end of NAD+ in the B-pocket of (c) Sir2Af2 chain A, (d) the template induced fit docked NAD+ of SIRT3 (3GLT). NAD+ in green and protein backbone in aqua. Three pairs of the residues that appear within 5 Å of the NAM end of NAD+ in both the Sir2 and SIRT3 structures are indicated as yellow arrow - F35 (Sir2)/F127 (SIRT3); red arrow - V195 (Sir2)/E323 (SIRT3); blue arrow - 196 (Sir2)/V324(SIRT3).
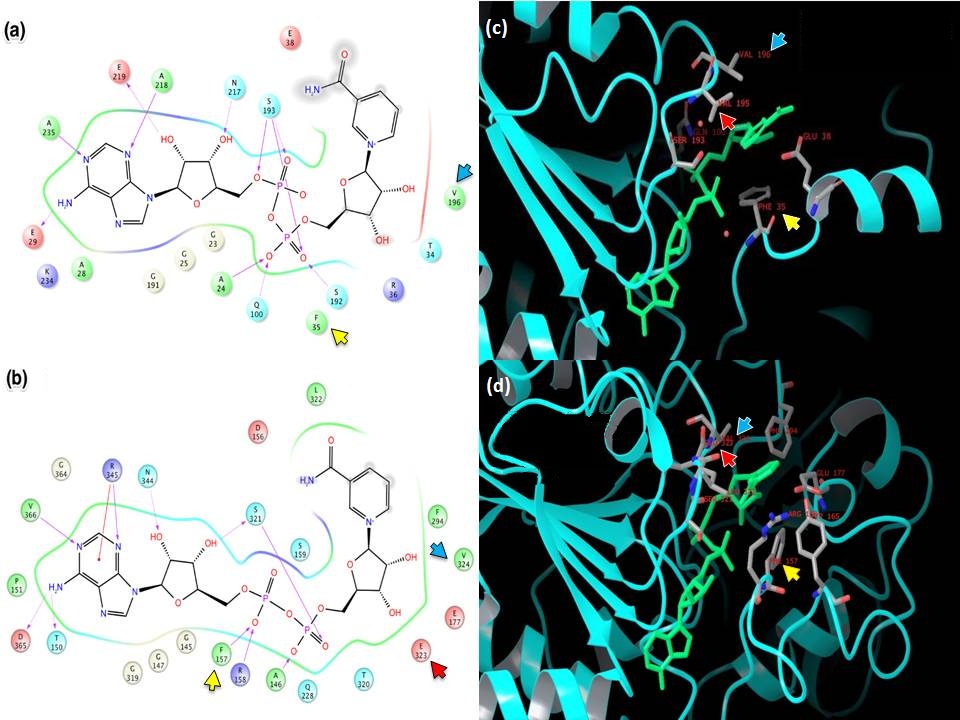
- Figure caption for the following figure: Figure XX. Multiple Sequence Alignment of Sirtuins with B-pocket Residues Highlighted. Yellow highlighting indicates those residues within 5.0 Å of the nicotinamide end of NAD+ induced fit template docked into the AB pocket of SIRT3. Red highlighting indicates those residues within 5.0 Å of the nicotinamide end of NAD+ in the 1YC2:A cocrystallized structure of Sir2. Orange highlighting indicates those residues that appear within 5.0 Å of the nicotinamide end of NAD+ in both the Sir2 and SIRT3 structures.

- Multiple Sequence Alignment of B-pocket residues that template induced fit moved.
- Task about how binding affinities are predicted using a training set of experimental data, with either LIA and/or MM-GBSA methods.
- Original task described as: 'Ok, in that case please point me to the relevant methods sections of the papers that you followed in carrying out these regressions, so I can summarize the methods. Are those papers on the wiki? I assume that there were no verbal notes on this in the notes section of the ppt."
- References to relevant papers:
- Tounge, B. A. & Reynolds, C. H. (2003). Calculation of the binding affinity of beta-secretase inhibitors using the linear interaction energy method. J Med Chem 46, 2074-82.
- An example of using Schrodinger's LIA (Linear Interaction Approximation; similar to LIE) in the Liaison program to calculate binding affinity.
- Aqvist, J. & Marelius, J. (2001). The linear interaction energy method for predicting ligand binding free energies. Comb Chem High Throughput Screen4, 613-26.
- One of the original papers on LIE (linear interaction energy)
- Liaison, version 5.8; Strike, version 2.1, Schrödinger, LLC, New York, NY, 2012.
- Two Schrodinger programs for calculating LIA and related linear regressions. In particular, read the introduction of the Liaison manual. The Strike statistical analysis package is used for the fitting and prediction analysis tasks of Liaison. For more information, see the Strike User Manual.
- Tounge, B. A. & Reynolds, C. H. (2003). Calculation of the binding affinity of beta-secretase inhibitors using the linear interaction energy method. J Med Chem 46, 2074-82.
- Task from RC (5-23): I'd like to confirm that Karthik has received a detailed protocol describing how the MCMM calculations were done, and that the earlier drafts of the paper contain all the relevant MCMM results; we wouldn' t want to lose all this work that was done.
- The MCMM method is based on Guimarães, C. R. W. & Cardozo, M. (2008). MM-GB/SA Rescoring of Docking Poses in Structure-Based Lead Optimization. J. Chem. Inf. Model.
- ToDo: add detailed protocol of how I used this MCMM method.
EK(6-18): Task 5, Figure: similarities/differences between (a) NAD+ template induced fit into the AB pocket of SIRT3 (3GLT) and (b) NAD+ cocrystallized in the AB pocket of Sir2 (1YC2 chain A). In this flattened 2D representation, residues within 2.8 Å of the NAD+ are represented as colored spheres, where: red=acidic, green=hydrophobic, blue=polar, light gray=(Gly). Solid pink lines are H-bonds to the protein backbone; dotted pink are H-bonds to the side chains. Solvent exposed ligand atoms are shaded gray.
Publication ready high resolution images are in the 'Pages and Files' in this wiki under the following names:
(a) 1YC2_A_NAD_cocrystallized_in_AB_pocket_of_Sir2_highlighting_residues_within_2.80_A_of_ligand.jpeg
(b) SIRT3_template_induced_fit_docking_into_3GLT_ligand_interaction_diagram_NAD_in_AB_pocket_.jpeg


Below table shows the sequence aligned residues that are in the above ligand interaction diagrams. Based on a ClustalW2 multiple sequence alignment of SIRT1-7 and Sir2Tm and Sir2Af2, highly conserved residues are in red (also marked by '*' in lower row), conserved residues in green (also marked by ':' or '.'). Grey highlighting shows corresponding residues that were missing within 3.0 A of the ligand.
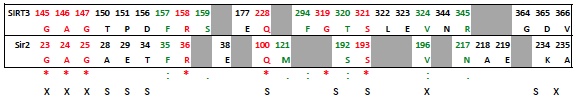
Residues that make similar interactions in both Sir2 and SIRT3:
- See above table. Residues labeled with 'X' in the last row have almost identical interactions with the NAD+ when comparing the SIRT3 induced fit structure to the Sir2 cocrystallized structure. Residues labeled 'S' are similar. A similar residue, for example, is when corresponding sequence aligned residues are in the same place in the Sir2 and SIRT3 structures, but only one makes (or can make due to side chain differences) an H-bond with the NAD+.
- Sir2:A28 does not H-bond to the purine amine, while the SIRT3 Threonine150 can with it's hydroxyl side chain.
- SIRT3:P151 does not interact with the amine on the purine because proline does not have a side chain to make an H-bond with and amine, where as Sir2:E29 does make an H-bond contact with the amine because Glutamic acid has a charged carboxylate.
- SIRT3:Leu322 covers part of the nicotinamide, which is solvent exposed in the Sir2 crystal structure (i.e., the equivalent Sir2:L194 is not within 3 A of the NAD+). The nicotinamide is more solvent exposed in the crystal structure of Sir2, while the induced fit structure in SIRT3 may have made more protein-ligand contacts in this solvent exposed area as an artifact of the induced fit docking process (which does not have explicit water). A similar case can be made for SIRT3:F294.
- Sir2:S193 H-bonds with 3 phosphate O, but SIRT3:S321 H-bonds with a side chain to the furan hydroxyl as well as a single phosphotidyl oxygen.
- SIRT3:R345 interacts with the adenine ring and the adenine amine, while the Sir2:Asn217 interacts with the the hydroxyl group on the furan ring. Note that the Arg is charged and the Asn is not charged, possibly creating this difference.
- Sir2:K234 (lysine) cannot H-bond to the adenine amine, but SIRT3:D365 can with the aspartic acid negatively charged side chain
Below is a full multiple sequence alignment of SIRT1 - SIRT7, as well as Sir2Tm and Sir2Af2 using the ClustalW2 algorithm. This sequence alignment is similar to the published multiple sequence alignment of SIRT1 - SIRT7 (Figure 2 in Jin, L., et al. (2009). Crystal Structures of Human SIRT3 Displaying Substrate-induced Conformational Changes. Journal of Biological Chemistry 284, 24394-24405). My alignment added the Sir2 sequences.
Multiple Sequence Alignment of Sirtuins.doc
EK (6-6): updating questions
- MCMM ensemble energy recalculated with MM-GBSA scoring function for NAD+: -107.90 kcal/mol
This uses the full 247 conformation of NAD+ that the standard MCMM found using MacroModel and its implicit water model. The 247 conformations are the lowest output conformations from a 10000 step macromodel conformational search. These 247 poses were imported into an MM-GBSA calculation with a dummy SIRT3 3GLT protein, where the dummy protein and the NAD+ conformations were > 20 A separated from each other, and where the MM-GBSA calculation did not minimize any atoms, except polar H-atoms. Thus, the NAD+ conformations remained < 0.3 A RMSD from the original MacroModel conformations. The "MM-GBSA Ligand Energy" was used in a standard ensemble correction, where <E> \= SUM_k( p_k * E_k ), where p_k is the partition function of the kth conformation:
p_k \= EXP(-E_k/(RT)) / SUM_i(EXP(-E_i/(RT))).- Macromodel project file: MCMM.prj job: mmgbsa_of_NAD_using_poses_from_mcmm
- Excel spreadsheet to get ensemble energy: MCMM_NAD_inH2O_10000steps.xls sheet:'MM-GBSA ensemble energy NAD'
- Excel spreadsheet with the updated ensemble energy correction with the MM-GBSA energies for Sir2 and SIRT3 AC/AB docking and in place scores. The updated MM-GBSA binding scores with the ensemble correction are in row 5.
EK (6-2): Answers to questions Raj texted Eric:
- In the cross docking with Sir2 in the AC pocket, do we really have a cocrystal structure with NAD?
Yes, it is a true cocrystallized structure with NAD+. PDB:1YC2 is a multimer with 4 separate structures of Sir2. One of them, from chain B, has NAD+ cocrystallized in the AC pocket. Chains A and D are two different instances with NAD+ cocrystallized in the AB pocket. - I don't see figure 7 referenced in the paper. Is that to be completely replaced by the new cross docking figure? And is fig 7 from the docking or the crystal structures?
The existing fig 7 in the paper is from the 1YC2 crystal structure. That figure is to be replaced by the cross docking figure. The cross docking figure has two pictures: (a) the cocrystallized structure from 1YC2 chain A compared to (b) the docked structure in the AB pocket. - Note also, for any validation figures, we need reported RMSD.
For the new figure 7 with the cross docking of 1YC2, the RMSD is 1.645 A (for non H-atoms) between the cross docked structure in (b in new figure 7 below) and the cocrystal structure from 1YC2:chain A in (a). There is another cocrystal structure with NAD+ in a slightly different conformation (the nicotinamide end in the B pocket is flipped) in 1YC2 chain D. The RMSD between the cross docked and this other AB pocket structure is better at 0.718 A. The ligand interaction diagram for the cocrystal structure (a in the new figure 7) uses chain A (with an RMSD of 1.645) because this is the structure that gives a lower energy, and is closer in energy to the in place score for the AC pose (from 1YC2 chain B). I guess I could have just as well created the ligand interaction diagram from chain D, which would have even more identical intermolecular protein-ligand interactions between the cocrystal and cross docked. Below is a picture of the superposition. (LEFT) RMSD 1.645 A. Blue is the cocrystal of NAD+ from 1YC2 chain A; orange is the cross docked. (RIGHT) RMSD 0.718 A. Red is the cocrystal of NAD+ from 1YC2 chain D; orange is the same cross docked.
a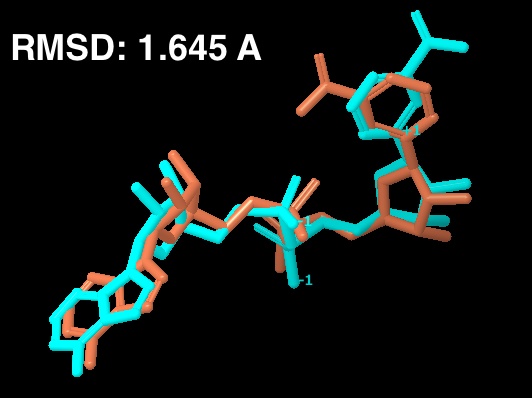
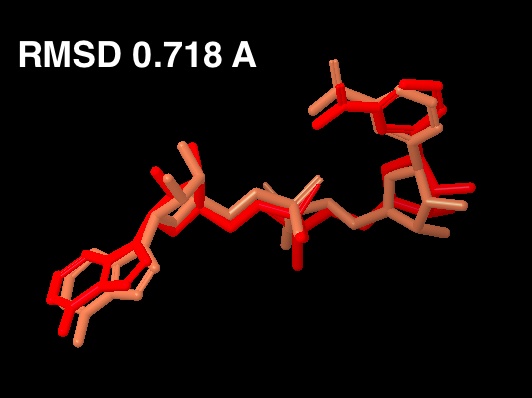
RC (6-2): I have edited the manuscript considerably in the computational results and discussion sections. I have accepted changes in the draft below in order to make these sections more readable, but I will use the earlier draft for further revisions of the other sections, which I have not touched. In this draft, I have organized the computational discussion section, and have copied the most important parts of our earlier correspondence directly into the newly organized section. My next step will be to use this correspondence to start writing the revised computational discussion section. Before I can do that, though , I need some more info.
Draft of JMB_060213C.docx
For Mon, in addition to the new figure Eric is working on, and his answers to the unanswered questions I previously asked, Eric needs to answer the following list of questions, which I excerpted from the revised draft above to save him time (a couple of these are related to this new figure). (Recall I indicated that I would have more questions during the process of paper revision.)
1) There needs to be a simplified table with the most important docking scores in the Results section. The current xls table is too big.
2) Results section - "Figure 5: The best docking starting structure for SIRT3 is 3GLT, which has the thio-intermediate of the acetyl-lysine peptide. The NAM has been cleaved and a bond to the thioacetyl is trapped. SIRT3, 3GLT with the trapped thio-acetyllysine ADPR intermediate. The B and C pockets are unoccupied because of the intermediate. H-bonds between the ADPR and the protein residues within 3.0 Åof the ligand are shown here."
Decide whether to mention similar results with 4FVT AC pocket here. Need RMSD between NAD docked to 3GLT AC pocket and 4FVT xtal.
- 1.53 Å : RMSD between NAD docked into 3GLT AC pocket (green molecule below) and 4FVT carba-NAD xtal (grey carbon atom molecule below)

3) THERE ARE NO FIGURES OF SIRT3 AB POCKET IN THE RESULTS SECTION; ONLY TABULATED RESULTS. DECIDE IF OK TO SHOW SIRT3 AB ONLY IN DISCUSSION
4) IN SIR2 AF1, IS IT NAD+ OR ANALOG IN THE AC POCKET? NEED RMSDS BETWEEN XTAL AND DOCKED FOR VALIDATION
ADD: INSERT SHORT PARA ON SIR2 CROSS DOCKING. CROSS DOCKING SHOWS ROBUSTNESS TO SMALL DIFFERENCES IN COORDINATES. FIRST CHECK WHETHER NAD+ OR ANALOG IS IN AC POCKET. THEN, SIRT3 AC POCKET DOCKING VALIDATED BY ABOVE TECHNIQUE (MORE RELEVANT TO 3GLT THAN 4FVT; 3GLT DOCKING COMPARISON TO 4FVT WOULD BE ANOTHER VALIDATION IN ITSELF. HERE, JUST MENTION THE RMSD.)
5) ADD: HOW 4FVT AC POCKET DOCKING CORROBORATES 3GLT RESULT (RMSD ONLY)
With the newer 4FVT structure, a figure showing the NAD+ induced fit docked into AC pocket of 3GLT compared to the cocrystallized structure of carba-NAD in the AC pocket in 4FVT would be another type of validation of the method. THIS WILL BE HANDLED BY REPORTING RMSD ALONE - see above
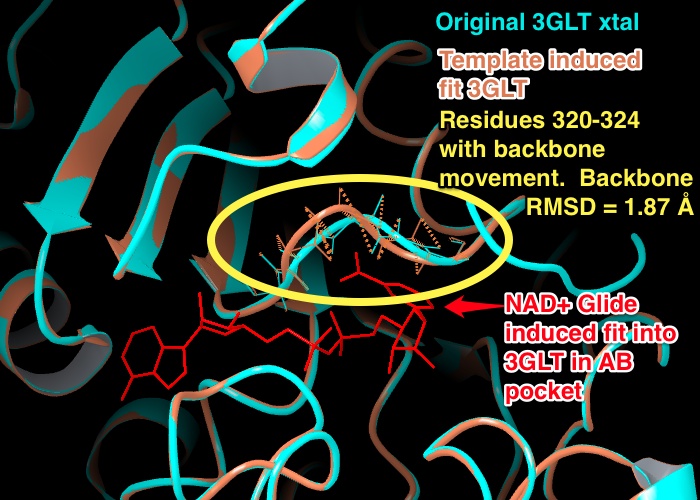
6) "The backbone moved only for residues 320 to 324, while the remaining backbone for other residues were either constrained or did not move." CAN WE PROVIDE A BACKBONE RMSD?
- EK(6-6): Backbone RMSD: 1.87 Å (non H-atoms of backbone) between residues 320 - 324 of the xtal structure of 3GLT vs. the template induced fit structure of 3GLT created to dock NAD+ into the AB pockets. Figure below.
7) INSERT AND DISCUSS ERIC’S NEW FIGURE SHOWING COMPARISON OF AB POCKETS FOR SIR2/SIRT3. IF HE HIGHLIGHTS SPECIFIC INTERACTIONS, MENTION THEM. MENTION RMSD, BUT DO NOT MENTION SCORES UNLESS WE CAN SHOW THESE INDIVIDUAL INTERACTION ENERGIES ARE SIMILAR IN MAGNITUDE IN SIR2/SIRT3 AB.
8) "Both Sir2 and hSIRT3 make similarly energetically favorable interactions in the AB pose, as well as in the AC [EK2] pose. The adenine and diphosphates have similar intermolecular interactions in the A pocket, especially with conserved residues. For example, conserved residues SER193 and SER321 form critical contacts with a phosphotidyl oxygen in Sir2 and SIRT3, respectively. As with the NAM in the C pocket cocrystallized structure of Sir2 and the docked structure in SIRT3, the carboxamide at the nicotinamide end of NAD+ in the AC binding mode makes a crucial hydrogen bond with Ile102 and Ile230 in Sir2 and SIRT3, respectively." CHECK IF THESE ARE HIGHLIGHTED IN ERIC’S NEW FIG
See above for answer to this task.
9) IN THE LAST 5 COLS OF NEW TABLE IN RESULTS. HOW DOES PROTEIN FLEXIBILITY ALLOWED IN TWO COLUMNS COMPARE TO THAT IN THE TEMPLATE INDUCED FIT ALGORITHM?
10) DESIRABLE TO SHOW A SEQUENCE ALIGNMENT OF SIR2/SIRT3 AROUND AB POCKET HIGHLIGHTING THE HOMOLOGOUS INTERACTIONS AND ALSO THE RESIDUES RESPONSIBLE FOR DIFFERENCES IN BACKBONE STRUCTURE.
See above for answer.
EK(5-28) to do list based on today's discussion with Raj:**
- Post updated table of MM-GBSA energy components with the fixed protein structure
- fixed protein structure had the overlapping H-atom minimized. NAD+ binding pockets remained frozen.
- include updated MCMM scores for Sir2 and SIRT3 AB/AC poses.
- updated spreadsheet below.
- MM-GBSA_components_Sir2_and_SIRT3.xls
- Corrected MM-GBSA summary from spreadsheet
| (kcal/mol) |
AB |
AC |
| SIRT3 MM-GBSA corrected 3GLT protein |
-82.4 |
-105.4 |
| SIRT3 MM-GBSA with MCMM for 3GLT |
-75.3 |
-95.2 |
- Figure: cross docking for Sir2. Side-by-side comparison of NAD+ cocrystallized in AB pose from 1YC2 with the cross docked NAD+ into the AB pose. The cross docked structure is the NAD+ docked into the Sir2 AB pocket starting with the Sir2 AC cocrystallized structure
- Figure X: Intermolecular protein-ligand interaction diagrams of NAD+ in the AB pockets of Sir2Af2 comparing (a) the cocrystallized structure from 1YC2, chain A, with (b) the cross docked structure. In this flattened 2D representation, residues within 2.8 Å of the NAD+ are represented as colored spheres, where: red=acidic, green=hydrophobic, blue=polar, light gray=(Gly). Solid pink lines are H-bonds to the protein backbone; dotted pink are H-bonds to the side chains. The cross docking method produced a pose with very similar intermolecular interactions as the cocrystallized structure. The lack of a protein "pocket" line around the NAM end and the grey spheres around those atoms indicate that the NAM end is exposed to solvent in the B pocket. In (a), there are no specific intermolecular interactions between the protein and the NAM end of NAD+. In (b), however, the NAM is slightly less solvent exposed in the B pocket of the cross docked structure due to the docking process lacking explicit waters in the B pocket.
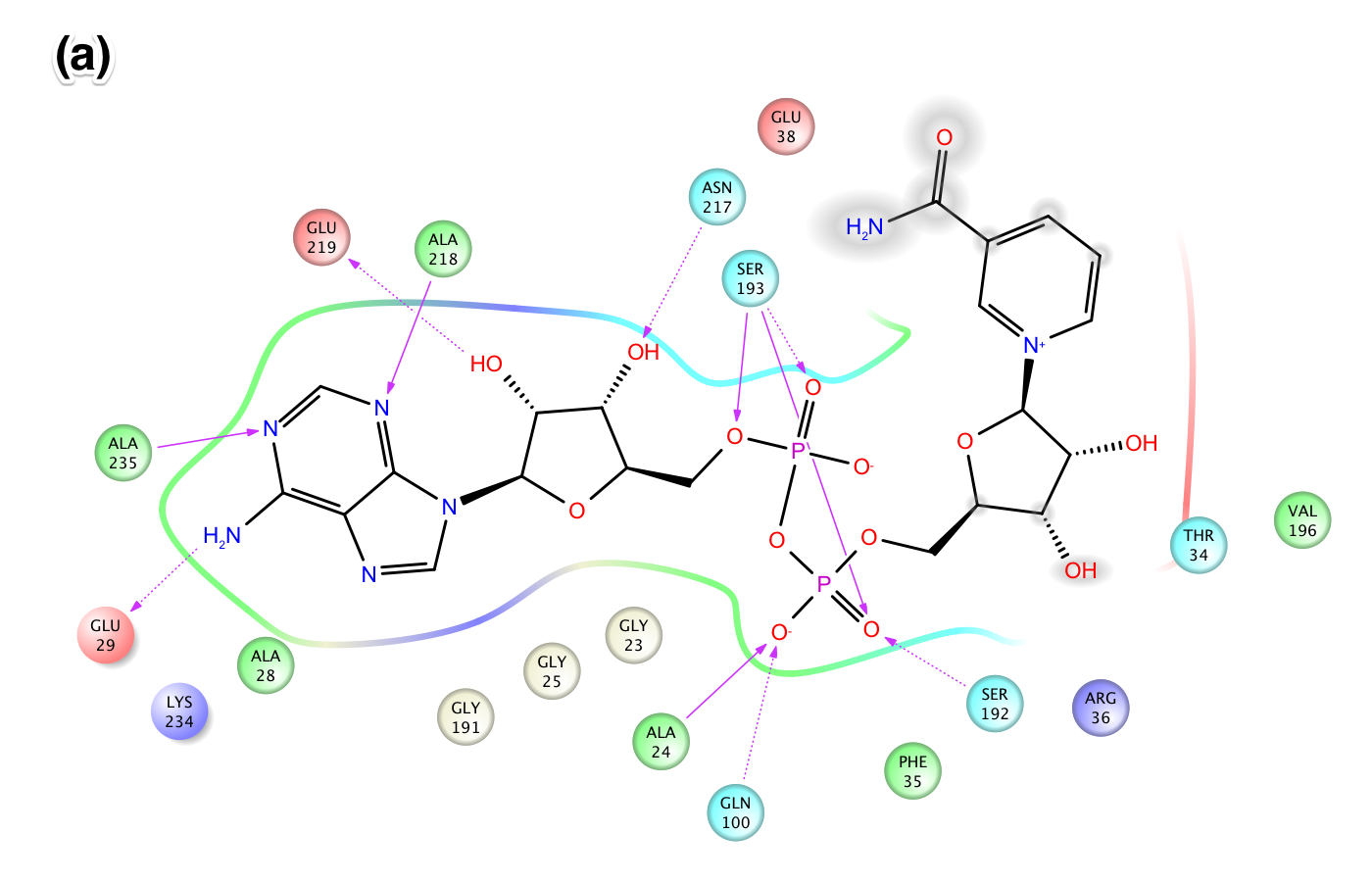
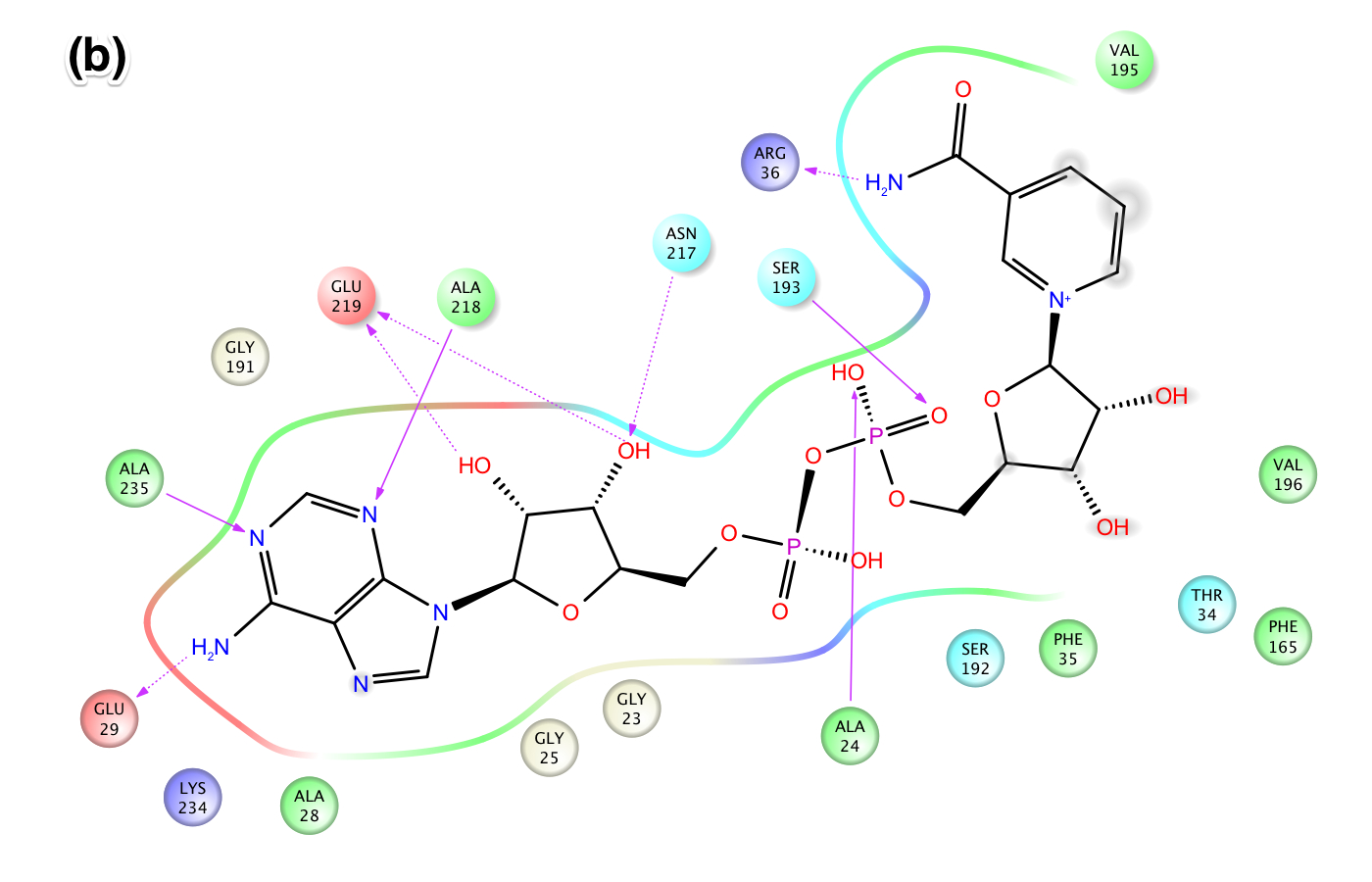
- Figure X: Intermolecular protein-ligand interaction diagrams of NAD+ in the AB pockets of Sir2Af2 comparing (a) the cocrystallized structure from 1YC2, chain A, with (b) the cross docked structure. In this flattened 2D representation, residues within 2.8 Å of the NAD+ are represented as colored spheres, where: red=acidic, green=hydrophobic, blue=polar, light gray=(Gly). Solid pink lines are H-bonds to the protein backbone; dotted pink are H-bonds to the side chains. The cross docking method produced a pose with very similar intermolecular interactions as the cocrystallized structure. The lack of a protein "pocket" line around the NAM end and the grey spheres around those atoms indicate that the NAM end is exposed to solvent in the B pocket. In (a), there are no specific intermolecular interactions between the protein and the NAM end of NAD+. In (b), however, the NAM is slightly less solvent exposed in the B pocket of the cross docked structure due to the docking process lacking explicit waters in the B pocket.
- Post the protein reorganization energy for SIRT3 3GLT for AB vs. AC NAD+ poses. Energies should be using the MM-GBSA solvation model, which can be found in the "receptor energy" output from the MM-GBSA calculation. The AB protein energy is from the template induced fit MM-GBSA calculations, and the AC protein energy is from the standard induced fit (NOT template) docking / MM-GBSA energy. All are from the 3GLT crystal structure.
- From last column of spreadsheet, the receptor energy for the crystal structure of 3GLT with NAD+ docked into the AC pocket with minimal minimization (to within 0.30 A RMSD of crystal structure coordinates) is: -11592.02 kcal/mol. This is 812.54 kcal/mol lower in energy than the template induced fit structure needed to dock NAD+ into the AB pocket (-10779.49 kcal/mol)
(4) and (5) only after above 3 finished. - 4FVT: dock NAD+ into the AB pocket of 4FVT. First try standard induced fit, then template induced fit.
- Figure: similarities/differences between NAD+ template induced fit into the AB pocket of SIRT3 (3GLT?) and NAD+ cocrystallized in the AB pocket of Sir2 (1YC2). Not sure we need this figure. The simplest measure of similarity is the RMSD between the NAD+ from SIRT3 and Sir2 backbone superimposed structures.
EK(5-28) Meeting schedule:
- Review of the latest postings: what do the new corrected energies show?
- Discussion of what we need to show in the discussion: validation vs. differences.
- I suggest we focus on validating the SIRT3 AB pocket score
- then focus on differences between SIRT3 AB pocket (from template induced fit into 3GLT or 4FVT) and Sir2 AB pocket (from cocrystallized structure in 1YC2)
- Discussion of figures: I think the relavent figures should be focused on points 2.1 and 2.2 above.
- Amend the timeline for remaining work:
- Post the full table of MM-GBSA energy components for the calculations redone with the fixed overlapping atoms from 3GLT template induced fit.
- Redo a more full calculation with this fixed structure - may not be necessary.
- Do a cross docking with 4FVT - do the template induced fit docking of NAD+ in the AB pocket of 4FVT (same as done with 3GLT). This may make more sense for our discussion - as it is a true cross docking, and, like with the Sir2 1YC2 cross docking studies, we could compare the cross docking of NAD+ in the AB pocket with the co-crystallized score in the AC pocket.
- Create figures: which ones depend on our discussion today.
EK(5-28) Correction: references to 3GLR below are supposed to be 3GLT. 3GLT is the structure with the trapped ADPR intermediate because of thioacetyl lysine substrate, while 3GLR is the hSIRT3 with only the actyllysine substrate. 3GLT was used for the template induced fit docking of NAD+. I switched the codes once and the switch then propogated. Now corrected below. Also needs to correct in paper draft.
Rest of updates being written now.
- Huge energies fixed: Regarding the spreadsheet posted below "MM-GBSA_components_Sir2_and_SIRT3.xls", I have corrected the unreasonable large positive energies associated with the protein van der Waals energy. I speculated below that this huge positive energy came from clashes of the non-minimized residues near the prime minimized residues of the binding site. This was not the case. The problem was a single overlapping H-atom with a C-atom on the far end of the protein no where near the binding pockets. Because these non-bonding atoms were within 0.7 A of each other, there was a huge positive energy penalty. I corrected this overlap through an energy minimization of that area of the protein, keeping the atoms within 6 A of the NAD+ binding pocket fixed, as well as the backbone atoms fixed, but allowed side chains to minimize in the problem area. This fixed that unusual energy and brought the protein van der Waals energy down to around -1300 kcal/mol, which is a reasonable value.
- Schrodinger software inconsistent: upon recalculation of the previous results with the overlapping protein atoms fixed, the results changed and the AB and AC conformations were closer in energy. This was not due to the correction of the overlapping protein atoms, but due to a change in the value of the free ligand energy. The row in the Excel spreadsheet labeled "Ligand Energy" (for the original calculations) shows -47.13 kcal/mol and -28.40 kcal/mol for the AB and AC conformations. With the new calculations, both the AB and AC ligand energy is around -48 kcal/mol. This actually makes more sense, because the free ligand energy should be the same for both the AB and AC conformations. It is unclear to me why the program originally had different values for the ligand energy. Interestingly, when I redid the calculations with the original protein structure with the overlapping atoms (same conditions as before), it had ligand energies of -48 for both the AC and AB. Thus, the software did not replicate its own results. I suspect this inconsistency arrises from a slightly different workflow. I can talk more about this, but it is an inconsistency. Regardless, the newer results make more sense - the free ligand energy should be the same for both the AB and AC.
- Corrected ligand and protein energies shrink the energy difference between AC and AB. The original values of -107.9 and -84.4 for the AC and AB MM-GBSA results, respectively, have a 23.5 kcal/mol energy difference. The newer results have about a 15 kcal/mol energy difference, still favoring the AC.
- Additional evidence supporting that AB docking is too high in energy in SIRT3. All of the results with the AB docking into SIRT3 3GLT do not use MM-GBSA with a flexible protein. The protein is prepared with the template induced fit method to move the sterically blocking side chains in the B pocket. Then standard Glide XP or SP docking is done, then MM-GBSA docking is done keeping the protein frozen. If additional induced fit MM-GBSA docking is allowed, the B pocket is re-closed and all docked conformations are in the AC pocket or something close to this AC pocket. This is what the protein rearrangement energy should show - the AB docked poses are in a more strained protein than the AC. I need to get these numbers from the newly relaxed protein structure.
- Still working on posting an updated spreadsheet.
EK(5-23) I am posting what I have so far today because I promised to update the wiki by 6pm. I still have to find and put numbers in the table (see notes), so, you might want to wait until tomorrow to look at this. Below is the breakdown of the MM-GBSA scores (all the components) for the Sir2 in place scores (AB and AC from the co-crystallized structures of 1YC2 chains A and B), the SIRT3 template induced fit scores (AB and AC pockets for NAD+ in 3GLT), and the newer in place scores for NAD+ in the AC pocket of 4FVT. Some notes also.
- MM-GBSA_components_Sir2_and_SIRT3.xls
- The template induced fit of NAD+ in the AB and AC pockets of 3GLT uses the same template induced fit structure. 3GLT was template fitted to accomodate the AB NAD+ pose, then this structure was separately induce fit docked to NAD+ in the AC and separately again the AB pose. The initial template induced fit to get the NAD+ into the AB pocket resulted in a huge positive Prime vdW energy, probably from clashes of the between non-minimized residues and residues within the prime minimized zone to accomodate the NAD+. I recall minimizing the rest of the protein to fix this, but I have to find the files. I will post later. The MM-GBSA dG binding still works because of the cancelation of errors, but that large energy for the vdW was troubling and I did address it.
- can you make any sense of the various components of the MM-GBSA energies?
- There is no breakdown of the components of each amino acid residue in this table. That can also be made if needed.
XG(5-22) The Table below is the summary of MMGBSA scores reported in current draft (not include new structure 4FVT). The Sir2 docking studies were started with PBD:1YC2. And old SIRT3 structure is 3GLT. Eric, please double check the cross docking scors of Sir2 for AC and AB pocket: the ones show at ACS presentation are opposite to the ones in the manuscript.
EK (5-22) The above table clarifies all the scores. Two comments: the SIRT3 (3GLT) scores of -107.9 and -84.4 are not "in-place scores"; they are the induced fit docked MM-GBSA scores. The two scores for SIRT3 (4FVT) are as close to an "in-place score" as possible with the transformed carba-NAD into NAD - so they can be considered in place scores. You can decide whether you want to report both of the 4FVT in place scores;
RC (5-23): Ok, so we have not redone the induced fit calculations in the AB pocket with 4FVT. That is ok, just confirming. See also my comments below - we would need to compare 3GLT AB pocket scores with 4FVT AC pocket scores. Which raises the question of the aim of including 4FVT scores in the paper. Is it only to validate those obtained with 3GLT?
EK(5-28) No, we have not done induced fit AB calculations with 4FVT. Although it might be a good idea. I could try induced fit calculations with multiple structures of SIRT3 - and, hopefully, if all of these structures result in higher energies for the AB template induced fit structure, this would fit with our hypothesis. Yes, the 4FVT scores do validate those obtained with 3GLT. It is unfortunate that we don't have a xray structure with NAD+ in the AB pocket in SIRT3 for a more direct comparison. The difficulty with our calculations that show that AB pocket binding is higher in energy than AC in SIRT3 is this: what if we are not sampling enough to accomodate the NAD+ correctly into the AB pocket? We are sampling a lot with the prime/plop algorithm, but really only within 7 A of the NAD+. These algorithms are very good at comprehensively sampling in this volume, especially for the side chains. But we are not exhaustively sampling.
EK (5-21): SIRT3 simulations results with new crystal structure:
- PDB: 4FVT SIRT3 with NAD+ analog (carba-NAD) in the AC pocket.
- Preparation Method:
- 4FVT protein prep with restrained minimization to within 0.30 A for heavy atoms
- co-crystallized carba-NAD transformed into NAD+ by replacing a CH2 with O on 5 membered sugar ring next to nicotinamide (this is the only difference between carba-NAD and NAD+)
- The complex of the SIRT3 crystal structure with the NAD+ (from the carba-NAD cocrystallized coordinates) is restrained minimized again (0.30 A RMSD for heavy atoms)
- In place MM-GBSA scoring of NAD+ in the AC pocket using the above prepared structure. Four MM-GBSA calculations done (mm-gbsa dG binding scores in kcal/mol) :
- -109.3 without cocrystallized H2O, and with no protein flexibility
- -81.4 without cocrystallized H2O, and WITH protein flexibility for both side chains and backbone within 5.0 A of NAD+
- -125.7 WITH cocrystallized H2O, and with no protein flexibility
- -96.3 WITH cocrystallized H2O, and WITH protein flexibility for both side chains and backbone within 5.0 A of NAD+
- Only SIRT3 AC pocket scores are shown because the only SIRT3 cocrystallized structure with NAD+ (or the NAD analog) has the ligand in the AC pocket. No AB pocket complexes are available.
- The above numbers are good results. They agree with the MM-GBSA dG binding scores for the docked structures into the other SIRT3 crystal structure from PDB 3GLT. Those scores are already in the paper with the top ranked AB pocket binding score of -84.4 kcal/mol and the top ranked AC pocket score of -107.9 kcal/mol. The number to compare the above results to is -107.9. The in place scores without protein flexibility and without cocrystallized H2O are about the same (-107.9 vs. -109.3). I believe that the -107.9 score from the induced fit docking was also done without crystallographic waters, but I have to check this.
- RC (5-23): Ok, please let us know.
- EK (5-28) this calculation is without crystallographic waters.
- Even so, when the crystallographic waters are included with the in place score for the new crystal structure, the energy is even more favorable at -125.7 kcal/mol. This even more energetically favorable result is what we would hope to see, because the premise of the paper is that NAD+ in the AC pocket is more energetically favorable than in the AB pocket for SIRT3.
- Including protein flexibility creates less energetically favorable MM-GBSA scores (-81.4 and -96.3) because the Prime minimization of the backbone and side chains within 5.0 A of the NAD+ results in a better Prime energy for the protein complex (the thing minimized). i.e., side chains are moved around to minimize their energy, but the binding energy happens to get less favorable because the minimization function is not based on the dG of binding score.
- I'm not sure if it is entirely fair to compare the previous induced fit docked / MM-GBSA SIRT3 AC pocket score (-107.9) to the in place scores with the new crystal structure without prime side chain and backbone minimization (-125.7 and -109.3). Why? because the -107.9 score is created with prime backbone and sidechain minimization, but the -125.7 and -109.3 scores do not have this prime minimization.
- RC (5-23): I agree, but this creates a small problem. If we do not report the 3GLT scores for the AC pocket, then how can we provide them for the AB pocket. Or do you suggest reporting them, but not comparing. See also my comments above.
- EK (5-28) yes report them but do not compare them.
- However, because the -125.7 and -109.3 scores come from the crystal structure coordinates, these scores are more reliable, and are the ones we should report.
RC (5-19): Further comments and information needed for 1st sirtuin paper revisions:
You can see why I was very confused by this writeup. Please provide your answers below my questions; I need some of these answers to complete my revisions. Please do not make any revisions directly in the paper - I will use the necessary info/clarifications you provide to make my edits. Thanks.
After my upcoming revisions, we may ask for the following additional info from Eric: a) final versions of the figures; b) docking results with the new SIRT3 structure if he has finished them and the results are conclusive; c) possibly, a ligand interaction diagram for SIRT3.
Eric's discussion sections:
“In addition to the investigation of NAM and isoNAM, protein-ligand docking using the newly available human SIRT3 crystal structuresfrom 2009 - 2013 52; 53agreed with previous crystal structures and provided insights into the differences...”
WHAT PREVIOUS CRYSTAL STRUCTURES? WHY ARE WE EMPHASIZING THIS IN THE FIRST SENTENCE? YOU MENTION DIFFERENCES BETWEEN SIR2 AND SIRT3 IN THIS SENTENCE, BUT THE REST OF THE PARAGRAPH SAYS NOTHING ABOUT DIFFERENCES. INSTEAD, ALL YOU TALK ABOUT ARE THE SIMILARITIES!
(EK) What I was trying to establish is that the docking poses for SIRT3 in the AC and AB pockets is similar to observed crystal structures with Sir2 and others, and not some different pose that would not be reasonable. There is a later paragraph that talks about the differences in the scores between AB and AC docking for SIRT3. I focused on the similarities of intermolecular contacts first to establish that the docking methods produced reasonable structures. There are also differences in intermolecular contacts that could be mentioned, but I did not because it is hard to figure out whether these difference are artifacts from the induced fit docking. It is also difficult to then attribute any one of these differences as the most important contributions to the difference in the overall docking score between the AB and AC docking.
The specific structures that this sentence refers to: SIRT3 is 3GLT and 4FVT; Sir2 is 1YC2 and 2H4F. Also, given the new SIRT3 in place scoring with the recent 4FVT structure, this paragraph can be changed to include that information, rather than only discussing induced fit docking / MM-GBSA scores into 3GLT.
RC (5-23): Regarding 4FVT, see my comments above. Regarding Sir2, when is 1YC2 referred to and when is 2H4F referred to?
EK(5-28) the paper refers specifically to 1YC2. 2H4F was used only as a reference, but is not in any of the figures or specifically in the text. The 2H4F is fo the Sir2Tm species, while 1YC2 is for Sir2Af2. 1YC2 is used because it has both the AC and AB poses for NAD+.
Overall, I think that this discussion section can be broken down better into 2 parts:
- Validation: why we should believe the SIRT3 results
- Cross docking with Sir2 works, so the method may also be good for SIRT3. RC (5-23): This part was not clear to me in the current writeup. I will revise it.
- Also add the similarity of the SIRT3 Carba-NAD crystal structure
- This is the parts you had many questions about - i.e., why are we talking about similarities, when we should be talking about differences between Sir2 and SIRT3. However, it is important to mention similarities, too, because of the high sequence homology, NAD+ should dock into SIRT3 with the same similarities observed in cocrystallized structures of SIRT3 with the carba-NAD analog or the ADPR intermediate. This validates that the docking poses for SIRT3 contain the same intermolecular contacts that are observed in Sir2 (1YC2) and SIRT3 (3GLT and 4FVT)
- RC (5-23): Are we talking about AC or AB pocket docking validation here, or both?
- EK (5-28) I was talking about validating AB and AC docking. However, the more important validation is for the AB cross docking - docking NAD+ into the AB pocket starting with the AC cocrystallized structure. While we have the AB cocrystallized "answer" for 1YC2, we don't have that for SIRT3. We only have the 4FVT NAD-analog in the AC pocket. So it is more important to talk about the validation for the AB cross docking.
- Cross docking with Sir2 works, so the method may also be good for SIRT3. RC (5-23): This part was not clear to me in the current writeup. I will revise it.
- Differences: why SIRT3 AC vs. AB is different than that of Sir2
- This gets trickier to explain, and I could use your help in understanding what is going on. The overall MM-GBSA dG binding scores for Sir2 AB vs. AC and SIRT3 AB vs. AC agree with the noncompetitive and competitive experimental results, respectively. However, I had difficulty understanding the individual components of those MM-GBSA scores. I think more investigation can be done here.
- RC (5-23): Ok, XG mentioned you will be posting these. This section is of course the most important part of the computational discussion.
- EK (5-28) I posted the Excel spreadsheet, and another updated one will be posted.
RC (5-23): I agree, we definitely need to distinguish between validation and differences, and the validation should be shorter - or, it should come after the emphasis on differences. Otherwise, the message gets lost, and it appears as if we are unsure of our results. Also, if validation comes first, we will need to indicate clearly upfront that we are discussing similarities in order to validate the method, and we will be talking about differences later. I will take care of all this editing based on answers to the questions above.
“Although the overall binding scores were different, the protein-ligand intermolecular interactions were similar for Sir2-NAD+ and SIRT3-NAD+, and the docked interactions agreed with cocrystallized x-ray structures of Sir2 and SIRT3” WHICH STRUCTURES? IN WHAT WAY WERE THE RESULTS NONTRIVIAL IF CRYSTAL STRUCTURES WERE KNOWN? NEED MORE INFO ON WHICH DOCKED STRUCTURES ARE BEING COMPARED TO WHICH X-RAY STRUCTURES
(EK) Yes, I was more specific here, and some additional clarification would also help. The rest of the paragraph names specific intermolecular contacts that are the same between the Sir2 (shown in Figure 7) and the SIRT3 docked structures. What is probably needed here is a different set of figures - figures that depict the similarities between the Sir2 crystal structures and the docked SIRT3 structures. Figure 7 is not enough. Figure 7 should be changed from only showing the ligand interaction diagram for Sir2, to a figure showing the AC pose for the Sir2 crystal structure compared to the pose for the AC docked SIRT3 structure. Similar H-bonds or intermolecular contacts would be highlighted. Another set of figures could be done for the AB pose. Even better would be to compare the carba-NAD+ in the AC pose of SIRT3 to the SIRT3 that was docked.
RC (5-23): I agree the figures would be useful - see also my comments below.
Here, again, the cocrystallized structures for Sir2 is 1YC2 and SIRT3 is 3GLT and 4FVT. With the new 4FVT crystal structure and the NAD+ analog in the AC pocket, we have in place scores available from co-crystallized structures for the following:
| Pockets --> |
AB |
AC |
| Sir2 |
1YC2 chain A |
1YC2 chain B |
| SIRT3 |
NONE |
4FVT |
RC (5-23): Ok, I believe the answer to this question is yes, since you have indicated that the induced fit poses have been validated.
My main point above is that whenever we refer to a docked structure comparison to a xtal structure, we need to either refer to a figure (as you mention above) or to specific intermolecular interactions - and if we do not have a figure, we need some way to classify the docked structures (e.g. by some naming system) so that when we refer to them, it is unambiguous which docked structure we are referring to. If we use figures, we could decrease the length of this validation discussion. I assume that in that case, you feel we could just refer to those figures without discussing the specific similar contacts one by one?
EK (5-28) sounds good to have a better naming system.
RC(5-23) Please let me know which method you prefer and if you are planning to make to figures, when you could make them. If it takes too long to make figures, I suggest we just use the approach of unambiguously referring to the structures and similar interactions as noted above. Can you list those here? I would like to have these for a back up plan in case the figures take too long.
EK(5-28) We could have 2 sets of figures for validation:
- validate the cross docked AB structure. one figure is the NAD+ docked into the AB pocket from the AC cocrystallized structure
- validate the docking of NAD+ into the AC pocket of SIRT3. compare NAD+ docked into 3GLT to the co-crystallized structure of 4FVT
RC(5-23) In any case, due to space limitations, let's bear in mind that we should not flood the paper with validation-related figures. Figures that are referred to in the context of validation should also be referred to when describing the differences between Sir2 and SIRT3. If we make new figures, ideally, we would have one that can be used to show the AC pocket similarities and one that can be used to show the AB pocket differences. Should I assume that none of the figures currently in the results section are pertinent to this discussion, since we do not refer to them? If so, please explain why - is it because you want to highlight similar interactions, and that would clutter the existing figures, or do we simply not have the right structures shown side-by-side? This is also important to know for my revisions.
EK(5-28) We need to talk about what figures you think are best. I suggested two sets of figures just above. The most important figure, though, would explain why hSIRT3 blocks the B pocket. Figure 8 kind of does this, and it could be improved. The question to answer is are there more bulky side chains in the B pocket of SIRT3 than Sir2?
“The similarity helps validate the induced fit docked poses of SIRT3, because the same protein-ligand contacts with conserved residues seen in the Sir2 crystal structures are observed in the SIRT3 docked structures as well”
THIS INTERRUPTS THE FLOW. ARE YOU TRYING TO SAY TWO THINGS SIMULTANEOUSLY – VALIDATION, AND SIMILARITY OF INTERACTIONS? IF YOUR PRIMARY POINT IN THIS PARAGRAPH IS VALIDATION, I WILL NEED TO STATE THAT UPFRONT, AND THE FIRST SENTENCE SHOULD SAY NOTHING ABOUT EXPLANATION OF THE DIFFERENCES BETWEEN SIR2 AND SIRT3.
(EK) similarity in intermolecular contacts is additional "validation" of the docked poses SIRT3. See the above proposed outline for this discussion section that has a validation section (to which this sentence belongs) and a subsequent "differences" section.
There are two validations: the cross docking for Sir2, and the fact that the poses for the docked SIRT3 structures are similar to the Sir2 structures. The SIRT3 structures are also different in some ways, but when I looked at the break down in the scores, it is difficult to attribute specific intermolecular contacts to the differences in score for SIRT3 AB vs. AC. Yes, the paragraph can be better phrased.
“Figure7 depicts the protein-ligand interaction diagram of Sir2Af2 cocrystallized with NAD+ in AB and AC pockets. Both Sir2 and hSIRT3 make similarly energetically favorable interactions in the AB pose, as well as in the AC pose”
ALTHOUGH YOUR COMMENT TO THE SIDE INDICATES THE PARAGRAPH IS ABOUT CROSS DOCKING VALIDATION, THIS PARAGRAPH SAYS NOTHING ABOUT SIR2 CROSS DOCKING! MAYBE IT WAS SAID IN THE RESULTS, BUT THAT NEEDS TO BE REPEATED BRIEFLY HERE.
(EK) Figure 7 would be better used to explain the Sir2 cross docking validation. In this paragraph I focused on how the SIRT3 docked poses were similar to the Sir2 crystal structures - so adding the cross-docked Sir2 into the mix would not be helpful in this paragraph. That comment was from a previous version of the paper and is not valid for this paragraph.
So Fig. 7 could be used with an additional paragraph (as outlined above) to talk about how cross-docking validates the methods. Specifically, Fig. 7 (which are from the cocrystallized structures of 1YC2) could be used to compare how similar the intermolecular contacts are between the crystal structure and the cross docked structure. I've done this - there are differences and similarities. The emphasis would be that the important intermolecular contacts are also made in the cross docked structures.
RC (5-23): Ok, do I have all the info needed to add this paragraph? Do you suggest referring to any particular contacts, or simply saying that the contacts are similar between the cross docked and crystallographic structures?
Are we talking about the current Figure 7, without any revisions?
EK (5-28) I think that figure 7 should be revised just to validate the AB cross docking. Side by side figures of a) figure is the NAD+ docked into the AB pocket from the AC cocrystallized structure b) the cocrystallized structure of NAD+ in the AB pocket. We should talk about what we're trying to do here.
Another figure is needed to compare the similarities between crystal structure Sir2 (1YC2 chains A and B) and the docked SIRT3 structures (docked into AB and AC pockets in 3GLT). But, with the newer 4FVT structure, a figure showing the NAD+ induced fit docked into AC pocket of 3GLT compared to the cocrystallized structure of carba-NAD in the AC pocket in 4FVT would be another type of validation of the method.
Again, an important part is to validate the result of the AB templated based induced fit docked structure into 3GLT for which there is no cocrystallized structure. A figure might help with this - comparing AB cocrystallized structures of Sir2 to this docked AB structure in 3GLT. You can think about which figures would be most important in the paper, because we are limited in the number of figures.
RC (5-23): Yes, see my comments above. The AB pocket validation is most important. Are you suggesting to have one new figure for AB pocket validation and one new figure for AC pocket validation?( If we use 4FVT to validate AC pocket docking, we need to be careful in explaining why we are using 4FVT for AC but 3GLT for AB. See my earlier comments on that issue.)
EK(5-28) Yes, exactly. We need to talk about what to do here.
"The adenine and diphosphates have similar intermolecular interactions in the A pocket, especially with conserved residues. For example, conserved residues SER193 and SER321 form critical contacts with a phosphotidyl oxygen in Sir2 and SIRT3, respectively. As with the NAM in the C pocket cocrystallized structure of Sir2 and the docked structure in SIRT3AND CARBA-NAD?, the carboxamide at the nicotinamide end of NAD+ in the AC binding mode makes a crucial hydrogen bond with Ile102 and Ile230 in Sir2 and SIRT3, respectively."
THIS IS POORLY WRITTEN PARTLY BECAUSE IT WAS NOT REVISED AFTER I INDICATED THE IMPORTANCE OF STERIC CLASHES AND PROTEIN REORGANIZATION. WHY DON’T YOU SAY ANYTHING ABOUT THE –DIFFERENCES—BETWEEN SIR2 AND SIRT3? HOW ABOUT STERIC CLASHES? THIS IS ONLY MENTIONED BELOW IN A SEPARATE PARAGRAPH? I WILL NEED TO OVERHAUL THIS PARAGRAPH. ALSO, I NEED INFO ON THE COMPARISON OF THE NAD+ AC POSE TO THE EXPERIMENTAL SIRT3/CARBA-NAD+ STRUCTURE.
(EK) again, this sentence is about the similarities as part of the validation. This sentence figures into the outline above. There can be additions added to talk more about the differences. Steric clashes of docking NAD+ in the AB pockets of SIRT3 3GLT are mentioned in later in the discussion section; the clashes are talked about in the context of why the template based induced fit method was needed. The steric clashes could also be used to talk about how the energy for the AB docking is less favorable, but a careful look at the components of the MM-GBSA score for those residues would be needed. Does the template based induced fit method does move these residues out of the way, but do they still contribute to less favorable energy?
There is a paragraph latter on about the protein reorganization energy.
RC (5-23): Yes, I saw that. I need to move that content up. However, the discussion is ambiguous about the causes for the differences in AB and AC pocket binding affinities. It does not clearly say whether the protein reorganization energy is primarily responsible for the differences in binding affinities. Importantly, although the binding scores are different even without the reorganization energy, there is no clear statement about where this difference is coming from. Especially given the amount of space dedicated to validation, the discussion of differences is particular short and seems like an afterthought.
EK(5-28) Understanding the difference in scores between the AB and AC pockets will come from an analysis of components of the updated MM-GBSA score spreadsheet. I will post and look at later, and could use your help understanding it.
"Predicted NAD+binding scores are similar for AB and AC binding modes for Sir2, but in SIRT3 the binding score for AC is lower than for AB binding. Although the cumulative scores are different in SIRT3, analysis of the individual energy terms…." HOW AM I SUPPOSED TO UNDERSTAND THIS WITHOUT THE NUMBERS HERE? PLEASE PUT THEM IN A TABLE. WHAT IS THE DIFFERENCE BETWEEN SUM OF INDIVIDUAL SCORES AND THE CUMULATIVE SCORE?
(EK) I am putting these numbers together for you in a large table. This table has the breakdown of components of the MM-GBSA scores for Sir2 in place scores and SIRT3 docked scores and the new SIRT3 4FVT in place scores. I will have this table for you tomorrow by 6pm.
RC (5-23): Ok thanks. I think the protein reorganization energy should also be included in the table. But before I look at that, can you clarify why there is an ambiguity - do you mean that the cumulative reported score is not the sum of the individual scores? That is the only way I can think of that the analysis of the individual terms does not explain the difference. This was one source of my confusion here.
EK(5-28) the cumulative score is the sum of the individual scores. i.e., coulomb, covalent, H-bond, vdW, etc... for each of the parts of the overall energy:
dG_complex - [ dG_freeLigand + dG_freeProtein ]
…to the binding score for MM-GBSA does not explain the difference between the Sir2 and SIRT3 scores, possibly because molecule’s large size.
"The largest part of the NAD+ molecule containing the adenine and the diphosphates is bound in a similar conformation in both the AB and AC modes for both Sir2 and SIRT3, masking individual energetic differences." WHAT IS THE MEANING OF MASKING AN INDIVIDUAL ENERGETIC DIFFERENCE?
(EK) I had a difficult time making sense of the break down of the MM-GBSA scores. I will provide the full break down of the scores for you and you can see if you can make sense of it. I could use your help in figuring out what to say here about the differences.
RC (5-23): Do you mean that the individual scores are dominated by contributions from the part of the molecule that is in a similar conformation?
EK(5-28) yes
"…. However, because the MM-GBSA scores do not include the full energetic protein reorganization penalty from the induced fit methods used with SIRT3, the less favorable AB binding in SIRT3 may be even more pronounced than reported here. The missing penalty" WHAT IS THE MISSING PENALTY? STATE IT HERE
(EK) the penalty is stated in the next sentence of > 100 kcal/mol.
SEE MY EARLIER COMMENTS REGARDING MOTIVATION FOR THE FOLLOWING PARA: I WILL NEED TO ADD IT.
"The NAM end of NAD+ adopts two flipped conformations in the crystal structures: one with the amide hydrogen of the NAM end of NAD+ in the AB pocket of the Sir2Af2 (1YC2 chain D) making an intramolecular hydrogen bond to the ligand phosphotidyl oxygen, and another with this amide pointed towards the solvent (1YC2 chain A). There are no hydrogen bonds in the B pocket with the outer half of the pocket exposed to solvent, allowing the NAM to move. This intramolecular H-bond is never seen in the docking, possibly because the energy is truly degenerate in this case where most of the NAM is exposed to solvent, or Glide has systematic bias against ligand intramolecular H-bonds. In addition, the NAM in the B pocket must move to the C pocket, and this degenerate flexibility in solvent could facilitate this motion."
(EK) you may want to leave out these sentences on the how Glide creates one pose with an intramolecular H-bond.
IF THERE WERE ANY PARAGRAPHS WRITTEN FOR THE PREVIOUS VERSION OF THE MANUSCRIPT THAT REFERRED TO HOW BINDING AFFINITIES ARE PREDICTED USING A TRAINING SET OF EXPERIMENTAL DATA, WITH EITHER LIA AND/OR MM-GBSA METHODS, PLEASE POST THAT IN A WORD DOC HERE, SO I CAN REUSE PARTS OF IT.
(EK) There was no paragraph written about this in the paper draft. This was presented in the ACS presentation; there are not extensive notes in the presentation about this; I verbally explained it during the presentation.
Here is the final ACS presentation:
Final_ACS_presentation_04062013.pptx
RC (5-23): Ok, in that case please point me to the relevant methods sections of the papers that you followed in carrying out these regressions, so I can summarize the methods. Are those papers on the wiki?
I assume that there were no verbal notes on this in the notes section of the ppt.
EK(6-18) Answer is above.
ALSO, PLEASE PROVIDE THE MCMM ENSEMBLE SCORES. I WOULD LIKE TO HAVE THE OPTION OF RETAINING THE MCMM PRESENTATION IN THIS PAPER. IF YOU SUGGEST OTHERWISE, PLEASE INDICATE WHY.
(EK) MCMM calculations resulted in conflicting numbers that did not agree with our overall hypothesis. I can look up the specific numbers again and post them if you wish.
RC (5-23): I'd like to confirm that Karthik has received a detailed protocol describing how the MCMM calculations were done, and that the earlier drafts of the paper contain all the relevant MCMM results; we wouldn't want to lose all this work that was done.
Xiangying's sections:
"Defining the inhibition modality is important for making quantitative comparisons among inhibitors of sirtuins, and is necessary for calculating the enzyme-inhibitor dissociation constant, Ki, from the experimental assays. Ki and the related protein-inhibitor Gibbs free energy of binding provide a means of defining the energetic contributions of specific types of interactions between groups on the enzyme and functionalities on the compounds to the overall binding energy of interaction."
AS DISCUSSED, I WOULD LIKE SOME MORE DETAILS ON WHETHER THE Ki IS NEEDED TO OBTAIN BINDING FREE ENERGIES, GIVEN THAT ERIC APPEARED TO BE USING IC50's. CAN YOU GET THE Ki FROM THE IC50?
XG(5-20): Kd for inhibition (Ki) can be directly related to the free energy of binding to the specific enzyme form as DGbinding = RT ln(Kd). Therefore, rational comparisons of inhibitor affinity for a target enzyme are best made by comparing the dissociation constants for the varying inhibitors.
Affinity for a target enzyme is not the only one criterion used to judge the suitability of an inhibitor for use in human medicine. Another equal important criterion is selectivity of the compound for the target enzyme relative to other structurally or mechanistically related enzymes. Thus the affinity of a compound among a number of potential enzyme targets commonly will be compared. Then any meaningful comparison of inhibitor affinity among these enzymes should be based on Ki value.
By knowing inhibitor modality (competitive/uncompetitive/noncompetitive), one may predict if the inhibitor has tight binding to a certain enzyme. For example, one compound will bind much tighter to its target enzyme with competitive inhibition mode than with noncompetitive inhibition mode. The reflection of Ki value of the compound to a certain enzyme, is Ki (competitive) < Ki (noncompetitive).
We can calculate Ki values from IC50 values using equation for enzyme-substrate and target-ligand interactions by different inhibitory mechanisms.
EK(5-15)
Latest draft below. Corrected most of comments from Raj, and incorporated Xiangying's latest changes. The major issues were not addressed in this draft. I am available to answer any questions you have as you finish the draft.
Draft of JMB_051513.docx
XG (5-15) Eric, to save time, could you please make the following changes in your final draft? Thanks!
(1) NEW Title: Mechanism of inhibition of the human sirtuin deacetylase SIRT3: computational and experimental studies
(2) Introduction_ first paragraph: Change "Sir2 was required for lifespan extension by calorie restriction" into "Sirtuins, the highly conserved enzyme homologues of the yeast Sir2, have been implicated in aging and the regulation of metabolism and genome stability."
EK(5-15) OK. I will transfer all data you need and will be available for any questions you have as you finish the draft. I will need to read the final draft you create, so please forward that to me before you submit it. I'm correcting more things in the paper and will post the latest draft by 5pm, but there is not time to make any major changes.
RC (5-15): We've discussed this internally and come to the conclusion stated below under my previous update. As noted, I will take the latest draft starting today and finish it myself, asking for data when I need it. Please post your final updates by 5 pm today. I will take whatever is posted by 5 pm today and revise it for publication, without iteration. I will not use a draft submitted after 5 pm. Eric can finish the work with the newer cocrystallized structure if he likes, and I will determine with him whether/how to include it. That will be the last computational work - we have decided to phase out the computational part of the project after that, as I described below.
EK(5-15)
There might be a misunderstanding of what this draft is: a draft. It is by no means ready for publication. My task was to create a working draft of the paper that can be crafted into the publication after iterative feedback from Raj. A part of the process is for Raj to make the important comments and suggestions he did below. Raj has found a number of grammar mistakes and oddly written sentences, as expected in a 30+ page document. A fresh pair of eyes reading the paper is important to point these things out. There are a number of major issues to address before this paper is ready, some of which Raj commented on below. For example, Raj has given past feedback and direction to include a lot of material, and some sections of the paper and certain paragraphs need better integration. Also, we need to account for the newer cocrystallized structure of SIRT3 (4FVT) with the NAD+ analog in the AC pockets. We've worked very hard on this paper and are in the final stretch of this process.
I've addressed some of the comments (see blow).
Draft of JMB_051413.docx
RC (5-14):
I read through the 051013 draft yesterday.
My conclusion: this is one of the most poorly written scientific papers I have read in some time. I am concerned about putting my name on it.
Grammar is part of the problem, but by no means all of it. There is a breakdown of logic in several sections (especially in some of the newly written paragraphs), including seemingly contradictory statements, and an overall lack of clarity as to why certain paragraphs have been written at all. As someone who has been heavily in this work, my inability to understand many of the paragraphs is a source of serious concern, since it is quite unlikely that a reviewer or reader will understand them. The lack of cohesiveness suggests that the authors were not always focused on the writing process, were not closely collaborating, and were often forgetting the main point of a given subsection. The result is a paper whose overall message is garbled.
Although some issues may have been mitigated in the most recent updates, due to above and the significant delays, I have concluded that I will need to finish the paper myself after Eric finishes his final read below. After Eric's submission tomorrow, we will be phasing out the computational part of this project until later this year. In the meantime we will assess the need (if any) for computation in future papers and decide how those needs can be met. Eric, please also confirm that you have provided Karthik with copies of all the data. If I need any of this data during my editing of the paper, I will ask you for it.
Here is a partial list of sentences in the 051013 draft that I felt either needed editing or were contradictory/unclear in meaning. I am listing them here so I can refer to them while editing.
(greyed out comments have been fixed; additional comments in grey)
- - Sir2 was required for lifespan extension by calorie restriction (required when and by whom?)
- (XG 5-15) The sentence itself is confusing. It has been changed into "Sirtuins, the highly conserved enzyme homologues of the yeast Sir2, have been implicated in aging and the regulation of metabolism and genome stability." It isdifficult to address the relationship between Sir2 and longevity since the debate (does Sir2 extend lifespan in yeast?) is always there. Firstly, the basis for yeast aging is the recombination events within rDNA that release a single repeat in its circular form, since the extra-chromosomal rDNA circle (ERC) can exponentially accumulate and kill the cell. Sir2 regulates the rate of ERC creation and therefore the rate of yeast aging (Sinclair DA, Guarente L. Extrachromosomal rDNA circles--a cause of aging in yeast. Cell 1997;91:1033–42.) Then, in the late 1990s a study demonstrated that deletion of Sir2 shortens yeast lifespan and that Sir2 overexpression extends yeast lifespan.(Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 1999;13:2570–80.) However, a possible explanation of the mechanism by which Sir2 regulates yeast aging came after a study that revealed for the first time the true enzymatic activity of Sir2-a NAD+ dependent histone deacetylase. (Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000;403:795–800. ) Moreover, another study showed that deletion of Sir2 blocked the beneficial effects of dietary restriction (DR) on lifespan, which suggests that sirtuins were required for the DR-mediated increase in lifespan.(Lin SJ, Kaeberlein M, Andalis AA, et al. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 2002;418:344–8.)
- - Carbanmylase
- - mammals, seven (this not a sentence)
- - Switch order of clauses in "given that SIRT3" - SIRT3 is a double-edged sword, since...
- - Belongs to vitamin B3 - not clear what this means
- - Activity are achieved
- - Knowing the inhibition modality of a ligand ....conditions for simulational studies ...enzyme ligand complex - this sentence makes no sense, even with the recent edits
- (XG5-15) The setence has been changed to "Knowing the inhibition modality of a ligand is critical for setting up conditions for structural studies of the enzyme-compound complex." For example: a compound was uncompetitive inhibitor with respect to one of the substrates of the enzymatic reaction. If one crystallize the enzyme-inhibitor complex in the absence of the substrate, he would not get the correct cocrystal, since inhibitor binding requires the presence of the ES complex in this case.
- - Order of magnitude comparisons correlate well with a rank ordering - this sentence makes no sense - a comparison cannot "correlate well" with a rank ordering. Mathematically very imprecise language
- - The basic cell biology processes - what does this mean?
- - the extensive efforts...are needed - which extensive efforts?
- - stand for experimental findings - what does this mean?
- - which reveals a different strategy - how has a new strategy been "revealed"?
- - docking use xray
- - what is the difference been inhibition mode and mechanism in the sentence "In this paper,..." They are listed as two things, but are the same.
- - was used to mechanistically compare - very confusing sentence
- - can equally bind and wait for the noncompetitive inhibitor to leave - this is not scientific language. Equally bind?
- - Unlike Sir2, fewer...were available for SIRT3 - grammatically incorrect
- - requiring the use of docking to the closest structure to NAD+ - what does this mean?
- *(EK) already commented that this sentence must be changed anyways before publication because of the newer SIRT3 carbs-NAD cocrystallized structure
- - template-based not template based
- - compares the docked - confusing sentence
- - rmsd ranges from - should say the rmsd ranges from
- - In addition to NAD+, standard docking... grammatically incorrect
- - Ic5 values in for
- - While the an pocket is not equally favored
- - In particular, the carboxamide - what is point of this sentence?
- - downregulation of sirt3 - first say something about upregulation?
- (XG 5-15) We mainly focus on inhibition. It is OK not mention upregulation.
- - Defining the inhibitors of sirtuins, and is required
- - Similar between; changed to "similar for"
- - Protein-ligand intramolecular interactions - intra or inter? Same expected protein-ligand contacts - what is an expected contact?
- changed to "inter"
eliminated "same expected" to "the same protein-ligand contacts with conserved residues seen in the Sir2 crystal structures are observed in the SIRT3 docked structures as well
- changed to "inter"
- - In addition to the investigation of NAM, protein-ligand docking methods agreed - how can a method agree? I have no idea what is being said in this paragraph
- "method" changed to "result"; This paragraph is about how the cross docking works for Sir2. Previous paragraphs are about the in-place scoring for Sir2, but we don’t have in-place scoring available for SIRT3, only a method more like cross-docking, where a different cocyrstallized structure was used. It is not a contradiction to say that the Sir2 and SIRT3 make similarly energicaclly favorable contacts – it’s important to point out how they are similar, giving validity to the docking. SIRT3 and Sir2 are still different in overall energy.
- - Apparent contradiction in message in the two paragraphs below:
- Sir2/SIRT3 make similar energetically favorable interactions in the AB pose as well as in the AC pose (grammatically incorrect as well).
- see above comment; it is important to establish that the docking poses were similar for most of the large NAD molecule. Most contacts are similar and similarly energetically favorable, The differences in energy is not from these similarities.
- Then, later - binding score for AC pocket is favored for SIRT3 (grammatically incorrect as well, since a score cannot be favored)
- changed to "binding score for AC is lower than for AB binding in SIRT3"
- - Apparent contradiction in message:
- Individual energy contributions do not explain differences
- Added this sentence pharase: "Although the cumulative scores are different in SIRT3, ..."
- Then, later - there is an energy difference
- So this is not a contradiction.
- Individual energy contributions do not explain differences
- - Similar intramolecular interactions with residues - this is wrong
- changed to "intermolecular" A few instances of "intra" were changed to "inter"
- - There are no references to a table with scores in the discussion, making it impossible to read
- yes, that would be a good thing to add.
- - There is no motivation for the discussion of the "NAM end of NAD"
- That was put in there because of your previous request to explain the differences between AC and AB poses with respect to the A vs. C pockets where the NAM end of NAD resides. It could be better integrated.
- - Fast, first estimation - unscientific language
- changed to "computationally tractable"
- - Mechanism provides insight for further research - vague
- - Inhibition relief compounds - is this a technical term?
- - Although requested 3-4 times, at the end of the paper there is no discussion of inhibitor binding affinity prediction techniques based on correlations between experimental and computational binding free energies
EK(5-14)
- Latest revision below. The following was fixed:
- consistent technical terms
- combining figures 7 & 8 and redoing those figures from scratch in high resolution and with larger fonts
- fixed more grammar
- Still to fix/do:
- Microsoft Word creates spacing errors when Xiangying and I open and save the versions between our two different versions of Word - one on Windows, the other on Mac. I believe it is a software error related to the review and changes tracking. This error is very annoying and makes the paper appear like it is riddled with grammatical errors, all of which we already corrected. I will post another version later tonight with this spacing problem fixed.
- One final read through the entire paper.
- Draft of JMB_051313_EK_XG.docx
EK(5-10)
- Latest revision of paper below. Fixed major issues including:
- grammar, discussion, final paragraphs, flow of paper, addressed Raj's comments
- Still have minor fixes to add, such as:
- consistent spelling of technical terms (i.e., co-crystallized vs. cocrystallized) There are a dozen or more to look at.
- combining figures 7 & 8 into one figure, and redoing those figures in higher resolution and with larger fonts / single letter amino acid symbols. Redoing this figure is necessary because of the requirements for figures in the JMB journal.
- Other issues to discuss with Raj:
- Move some of the very long computational methods section to a supplemental section
- possible more discussion of NAM and isoNAM docking. There already is discussion of this, but do we need more?
- Important: how to include the latest cocrystallized structure of the NAD+ analog (carba-NAD) in hSIRT3 AC pockets.
- The 98% of the work for the paper has been completed. The above mentioned next steps are some of the little details to work out for publication formatting requirements, and other minor fixes. Raj can read this version; another version with more minor corrections is forthcoming.
- Draft of JMB_051013_EK.docx
EK(5-9)
- Discussed with Xiangying the the latest version of the paper.
- Tomorrow, Xiangying and I will finalize the draft by fixing the figures and editing the last two paragraphs of the discussion.
- we'll post the final version tomorrow by 5pm.
- Latest version:
- JPEG files for my figures are here: Dropbox:PMC-AT Research/Eric/Paper_images/
- most images are there, except the ligand interaction diagrams.
- Xiangying, are the resolutions high enough for publication?
The combined the figures from 1-8 have been prepared as pdf file. Eric please check if you agree with it. Plus the resolution of Figure 7 a and b are not good. Please send me the image files.JMB_05102013 figures.pdf
EK(5-2) updated plan:
- Final revised draft of paper with all my work by Tuesday noon.
- If I have time, I can revise the entire paper, but only after I've completed the updates to my sections.
- Add sentances about the energetic cost to use induced fit - protein re-organization energy. MM-GBSA does not incorporate this into the ∆G_bind approximation. This is an additional energetic cost not included in the MM-GBSA estimate. It is the PLOP or PRIME reorganization energy with SGB solvation model. List this as a separate energy that makes the existing MM-GBSA numbers look even worse.
RC (5-3): Thanks Eric. Don' t wait for the last minute to finish your work, and try working during the day. If you work consistently starting now, you should have plenty of time to finish all your sections - including figures and references - by Tues noon.
EK(5-2) talk with David Churchill grad students at Kaist
- I talked with Yonghwang Ha <[email protected]> and Dhiraj <[email protected]>
- gave them April ACS presentation about SIRT3
- Looking for collaboration to use PET, FRET, or ICT to measure protein-ligand binding distances and kinetics for inhibitors, activators, or allosteric activators using fluorescent dyes they can synthesize and tether to a protein or small molecule. They also suggested using the fluorophore directly as the ligand.
- Up to Raj and David to work out what's next
- Also possibility to have them synthesize drugs we computationally screen, but this may be better done with a contract specialty chemical company because of speed and IP issues.
- How to design a clever fluorophore experiment to find out something about sirtuins.
EK(5-2) After working hard for 2 days on the paper, I have made a lot of progress, and the final draft will take longer. Let me continue to work on the paper without Xiangying or you editing the below posted draft. Much time was spent analyzing the ligand interaction diagrams, as well as protein-ligand docking poses to fill in the important discussion section about why NAD+ binds with the same energy for Sir2, but very different energy for SIRT3 in the AB vs. AC pose. It is difficult to understand why because there are so many energy components in the MM-GBSA score, and a cancelation of terms. So I decided to explain the difference between Sir2 vs. SIRT3 AB/AC scores as cumulative rather than from any one particular difference. It is always possible to come up with plausible individual effects, but these individual effects get drowned out by larger numbers in other MM-GBSA terms.
Other things updated are additional details about the NAM and isoNAM docked poses, important because the laboratory emphasis on these molecules. Xiangying asked if I included binding affinity of NAM or isoNAM. Yes, I did calculate the score for NAM binding using MM-GBSA, but I did not include this in the paper because the score is not a good measure of the binding affinity of NAM due to base exchange with NAM. Reporting the number could be confusing. It is more important to discuss that docking resulted in a similar binding pose found in the x-ray structure of Sir2 co-crystallized with NAM.
Another question was about why I erased the MCMM ensemble correction section. MCMM was not used in the MM-GBSA calculations.
MCMM was not used for comparing NAD+ AB vs. AC binding. A big part of the correction for MCMM is more accurately calculating the energy of the free ligand (using an ensemble of low energy free ligand conformations). But it does not matter as much for comparing the different NAD+ AB vs. AC poses because the free NAD+ ligand energy is similar in both cases. So the correction error would mostly cancel out. MCMM would still theoretically correct for some differences. Standard MM-GBSA calculates the free ligand as a simple energy minimization of the bound state. Since the AB and AC poses are different, the free ligand energy is different for the different poses. While technically incorrect, there might be some cancelation of errors. While the MM-GBSA numbers could theoretically be improved, MCMM is not as widely used as standard MM-GBSA. And the MM-GBSA numbers agree with experiment and our hypothesis. If we started correcting with MCMM, we should also start correcting for many other known limitations of the MM-GBSA approach, such as a single protein-ligand state, as well as the lack of a more comprehensive sampling technique to find the lowest energy states for the AB and AC poses. What we have works within the limitations of this MM-GBSA method.
Please let me continue to work diligently on this draft. I will post another update tomorrow morning. I think the best plan is let me continue to revise the draft without you updating the draft. I don't think you should even read this draft other than out of curiosity to see the progress.
XG (5-2): Thanks, Dr. Raj, for editing experimental parts. I will modify the paper based on his comments. In the latste draft, a message of "[[file:/C:\Users\xguan\Documents\AppData\Local\Microsoft\Windows\Temporary%20Internet%20Files\Local%20Settings\Temporary%20Internet%20Files\Content.IE5\9N1O72K2\Documents%20and%20SettingspmclabMy%20Documentsxg%22%20l|../../../AppData/Local/Microsoft/Windows/Temporary]]" was found in the 2nd paragraph of Introduction. Can anyone explain why?
Eric ?RC (5-1): I have gone through all of XG's sections in her latest draft below and made detailed comments and edits throughout. I am now waiting on Eric's final draft Wed night; which will need to be merged with this after he posts it. Eric, please don't make us ask again for this.
XG, you should stop doing lab experiments for the next couple of days - I noticed that your sections were not carefully proofread or edited. You need to devote full time to answering my questions and finalizing your sections. There is also some work you need to do jointly with Eric, as discussed.Draft of JMB_050113_RC.doc
XG(4-30) Disucssed with Dr Raj and add the more comments on the draft. Draft of JMB_043013_PM.doc Eric: (1) you mentioned that your FINAL draft will be ready on Thursday morning. Dr. Raj need 2 days time to review and make change on the draft this week. Therefore, It will be appreciated if you can post it Wednesday night.
(2) Please response to ALL of the comments that Dr Raj has made.(3) Do you delect the contents of MCMM? You have calculated binding affinity of NAD+ docking into AB and AC pockets, have you ever done on NAM?
(4) I noticed that something was wrong with the references. Some of your references were missing in the very last draft. Please fix them or give them to me I can add them into EndNote.
(5) Thank you for editing Abstract and Introdcution. Please focus on and finish your sections.
EK(4-29) Another draft. Here, edits to the abstract and introduction. Fixed grammar, sentence structure and flow of paragraphs. The edits will take time for me to go through the paper line by line, including fixing grammar, ordering, and the requested revisions of the discussion. All of Raj's comments are being addressed. Work to continue on Tuesday.
Draft of JMB_043013_ek.docx
EK(4-29) Raj, I will continue to revise this draft.
This weekend I needed to redo some of the calculations for Sir2, to verify the cross docking and in place scoring results. When I searched through the original data, the results I found were not consistent with the results in the paper for one of the in-place scoring. I knew that I had previously calculated a different number that agreed with the rest of our results, but I could not find it. It took me half a day to resolve this, which ate into time for editing the paper. In particular, there are two structures in 1YC2 of NAD+ cocrystallized in the AB pocket. The one from chain D resulted in a score of about -65 kcal/mol (not consistent with our hypothesis because AC in-place scoring would be largely different at -99.0 kcal/mol). A recalculation with Chain B (also AB co-crystallized) results in an energy (-95 kcal/mol) consistent with the hypothesis in the paper that NAD+ binds equally well in the AB and AC pockets of Sir2. Since the general method for choosing the structures is to use the best scoring (lowest energy), it is consistent to choose the -95 kcal/mol result. It is resolved.
RC (4-29): I'm seeing very few changes to the draft. The discussion section indicates "still needs work" and it appears the main change was to cut and paste paragraphs from the results and put them in the discussion section. There are spelling mistakes and grammatical issues (see for example, the last para of discussion, which also needs reordering of sentences), which indicates quick changes were made and that the paper will need multiple rounds of editing involving me. In addition, even old comments that I made a week ago are often not yet answered. I'll need to think carefully about next steps for this paper, as well as how feasible it will be for us to write a second paper, since I do not want to have unreasonable expectations for this project and the current iterative procedure for editing is not suitably efficient.
EK(4-29) Below is the updated draft that merged my and Raj's drafts.
Draft of JMB_042913_ek2.docx
EK(4-29) test to see if get email notification when update to wiki.
EK (4-29) Below is the latest draft with my revisions. Important: my browser did not properly refresh and I did not see Raj's JMB_042713_rc.docx draft until after I had finished my revisions. I will merge the two documents on Monday. So this draft may not reflect all of the comments and revisions in Raj's document. This draft incorporates most of the major changes Raj and I talked about on Friday.
RC (4-27): After discussion with Eric on Fri, I have gone through the Sections he indicated were roughly complete, answered his questions, and indicated additional required changes. This does not include the computational discussion section - here, I have only indicated what changes Eric and I agreed upon. I will need to look at it again after Eric's next posting. I have not carefully read the experimental parts of the results, discussion and method sections yet, but I have made some minor comments to those sections as well, for XG. In addition, I have gone carefully through the intro and abstract, and made multiple suggestions for changes.
Eric must post his remaining changes by Sunday night, so we can do our next iteration Mon am. After his update, I will also look more closely at the experimental sections. XG, please make your remaining changes to the experimental sections by Mon.
Starting Mon, Eric will finish any remaining figure prep/simulation tasks that remain on the table.
XG (4-25):
(1) Comments on Figure 6 (page 12): JMB requirements about composite figures is listed as following: No more than four sections should appear in a single figure. Label individual sections in composite figures clearly with lower case letters, using (a), (b), (c).
(2) Eric, I looked through the current draft and have a TO DO list below. We can discuss if the points raised up here are necessary and also please indicate the missing points:- Introduction_last paragraph: are you fine with the current form or you want to add some background info of docking and MMGBSA? Please also provide the references.
- Results_3rd paragraph: the cross-docked scores need to be added.
- Results_Figure 4 and 6: use (a) and (b) to label the left and right sections.
- Results_Figures 8 and 9 plus description: you suggested in your comments to move them to discussion section. Do you mean the Results section is ended at Figure 6.
- Results_last paragraph: You said that I don’t think the differences in the NAM and iso-NAM pose is important; it is only important the they docked to the C pocket. Are you going to delete the whole paragraph?
- Discussion: Based on responding Dr. Raj's comments, could you please list how much work need to be done and how long it will take?
- Methods: Can you summarize it and move the details to supplimental section?
Wed, Apr 24, 2013
- Edits were so extensive in parts, that the Microsoft Word "tracking changes" comments became confusing with so many changes. I did not track all specific changes, and highlighted the most important changes with comments.
- changed "binding estimates" to "scores" with an explanation in results section that
"While MM-GBSA scores, which are reported in kcal/mol units, are not absolute binding affinities, they have been shown to have a good correlation to experimental binding affinities for many protein-ligand data sets" - planned major changes are in comments.
RC (4-24): This document is getting very hard to read. It has an assortment of colors and I cannot tell which edits were the most recent from Eric. Also, Eric has not provided any comments regarding his planned additional changes, as XG and I have started to do. Eric must post again more clearly by the end of the working day EST today (i.e., by 6 pm). Eric's schedule for changes is completely unclear at the moment, and we are concerned.
Tues., Apr. 23, 2013
Latest paper update. Still to edit much more.
Draft of JMB_042313.docx
Thur., Apr. 18, 2013
Some of the results for SIRT3 using the newer PDB:4FVT crystal structure.
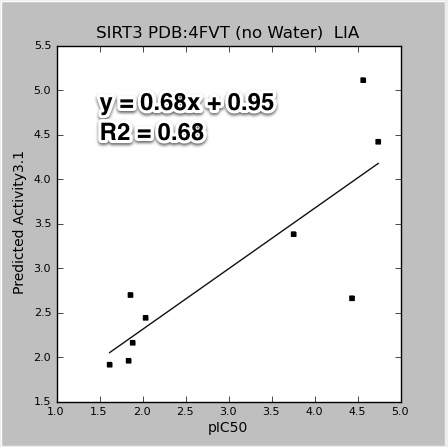
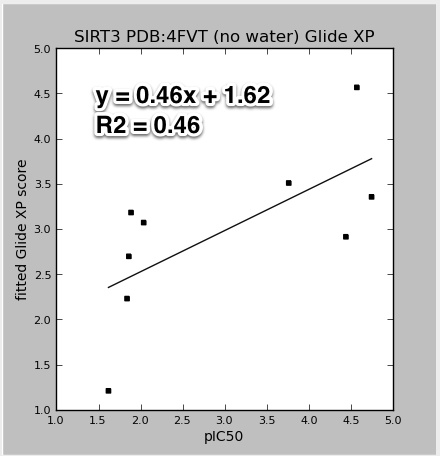
The data points for the above figures are for all the SIRT3 inhibitors: AC93253, Salermide, EX-527, 1-methylnicotinamide, nicotinic acid N-oxide, iso-nicotinamide, nicotinic acid, pyridine N-oxide, and nicotinamide. The results would improve with the nicotinamide outlier (at pIC50 4.4) left out. Also, all the models do an impressive job accounting for the wide range of activities. Except for nicotinamide, all the smaller single ring molecules have much lower inhibition in the mM range. It is not clear to me if these good results would hold up with decoy molecules (molecules that are similar to potent inhibitors, but are known experimentally to not bind).
Also note that the Y-axis in the above plots are not the raw GlideXP, or MM-GBSA scores. The linear regression fits and scales those scores to correspond to the same range as the pIC50. Plots for the publication should not have this scaling. I will fix.
(EK 4-4) Latest power point here:
Dropbox/pmc-at research/Eric/ACS_slides_04.03.2013.pptx
RC (4-1): My comments on slides and paper below (in no particular order).
(EK 4-2) Eric's comments from 2013.04.02.- Regarding LIA, this is one example of what I was referring under "other methods" below (see EK's notes from out meeting); i.e., a parametric model that is estimated for a particular protein/congeneric series based on fitting binding affinity rather than docking pose. The basic idea is to develop a model tailored to a specific system rather than relying on MM-GBSA correlations alone. EK, recall I mentioned that I would like Karthik to be involved with this going forward. See my comments below.
- (EK 4-2) Yes, I will talk with Karthik at ACS about this. LIA is one type of model parameterized to the particular protein binding site like you imagined.
- I saved a slightly edited ppt in dropbox
- LIA: there is a risk of overfitting when you use models with more parameters. This is because it is easier to fit a model with more parameters to a dataset. Usually, one gets around this problem by reported adjusted R^2's. EK, see below - ask KM to work on this given your data.
- (EK 4-2) Yes, part of the core of LIA is a 3 parameter multiple linear regression to the experimental ∆G_bind or pIC50. I could not use LIA with the small data set (of strong inhibitors) we have so far with SIRT3 inhibitors. As we get more data ( > 5 or 6) we can begin to use this. But at 5 data points to regression parameters, there is a risk of over fitting. A more comfortable data set size is >8, which is what is available with the SIRT2 data set I used with 20 molecules. The LIA method worked very well and is not over fit in this case.
- LIA: what type of sampling is used to compute the expectations and are these all done by the Liasion software under the hood
- (EK 4-2) The input into the LIA method (Schrodinger Liaison protocol) is the set of best docked structures from GlideXP, with one docked pose per ligand. Liaison runs a molecular mechanics (MM) minimization (truncated newton minimizer with the SGB implicit water model) for the docked protein-ligand complex, and a separate minimization of the free ligand and apo protein. The three of the components of the minimized energy function for the free and docked complex are parameterized with a 3 parameter multiple linear regression against experimental data. The three components are van der Waals energy, electrostatic energy, and the cavity energy terms of the SGB continuum solvent model.
- Liaison is NOT sampling additional states in the same way that Induced Fit or Prime MM-GBSA does with PLOP under the hood.
- EK must add detailed notes below each simulation slide that can be used directly for a section of the paper
- Do not write up LIA methods in the paper until we determine whether these results will be put in this paper
- XG and EK: present the talk to KM later this week to check timing. Most likely, it will run very long. you will most likely need to eliminate some slides. i am ok with you making these decisions. I think it is clear which slides contain our results/methods (most important) and which relate to prior art (less important)
- Leave EK time to present his part (important) - may require faster presentation of experimental section.
- (EK 4-2) yes, because we have so much material to fit into 20 minutes, we will need to leave things out.
(EK 4-2) will add comments to below as I edit slides. I have not added comments to below yet.
Slide-by-slide comments: the following comments apply to the paper as well as talk
-I changed the title
-slide 5 - need to decide which parts of biology to emphasize. Could either emphasize liver application (inhibition), or could mention both inhibition and activation applications, suggesting that understanding of inhibition mechanism is important to design of both. Need to say something about how the peptides you are using relate to the protein substrates of SIRT3, minimally should say that the protein sequence does not greatly affect the inhibition kinetics.
- (XG 4-2): modified slide 5.slide 5.pptx
- (XG 4-2): move it for backup slide.
- (XG 4-2): May not mention if the time is limited. But will prepare for answering questions.
- (XG 4-2): To support, NAM has similare inhibition effects on Sir2/SIRT1, but SIRT3 by showing the sequence alignments of Sir2 vs SIRT1 and Sir2 vs. SIRT3. I am waiting for Eric's slide and not sure it will show any similarities/differences.
- (XG 4-2): OK
- (XG 4-2): will do.
-slide 23 - does not appear essential for talk (thought it should be mentioned in paper)
-slide 26 - i agree that for the paper we may redo the noncompetitive/competitive calculations with the Carba-NAD structure, but see my comments below on paper writing (any new simulations should occupy only 25% of total time).
-slide 28 - mention that if the MM-GBSA/LIA correlations are strong (e.g. like some of those for SIRT2), we will use this method to discover new inhibitors
-slide 30 - what do we mean by promiscuous?
- (EK 4-3) removed name; I meant non-conserved residues.
--The paper should be finished based on current content
--After the conference, XG and EK devote 75% time to finishing write-up of paper based on slides. EK's efforts will be esp important since his parts of the paper are largely unfinished.
--KM learns LIA and reports adjusted R^2's. (KM may help w discussion of LIA).
--Was LIA reported in the referenced SIRT2 paper? If not (unlikely), the results could be shown in this paper
- (EK 4-3) Yes, a type of linear integration method was used in one of the papers, but it is different in that it uses MD snapshots.
--If the paper revisions are done before the latest congeneric series data and sims are done, we submit the paper in that form. It may even be better to save the new results for another paper.
Future work:
--KM continues running LIA for follow-up paper if it is not finished for this paper
--KM looks at SIRT6 structures, considers application of LIA; XG does kinetics of inhibition and inhibitor screening only if the computational/structural analysis warrants it
--KM considers development of LIA models for protein design (proposes what experimental binding affinity assays with inhibitors would be needed to train the models)
--KM's goal in attending talk is to plan out this future work
--XG finishes continuous coupled assay (to be used in second paper) and considers protocol for base exchange kinetics
Mon, Apr 01, 2013
See comments below on Mar. 28, 2013 by EK
Thu, Mar 28, 2013
Notes from Mon. phone call with Raj and other notes on slide preparation:
- while preparing the slides, also think about continued preparation of the paper
- Slides to create:
- poses and analysis of tight binders of SIRT3, and how they compare to published analysis of SIRT2 or Sir2
- Slide "SIRT3 Inhibitors: Docking into 4FVT". superpositions and ligand interaction diagrams
- slide with graphs of Glide, MM-GBSA, induced fit, and LIA for the 9 inhibitors we have for SIRT3. Show correlation. Discuss why some methods do not show correlation
- Slide: "Binding Energy Prediction: SIRT3 Inhibitors" We only have data for 3 applicable inhibitors of SIRT3 for this: EX527, Salermide, AC93253. The lower affinity mM small inhibitors do not work well in building a binding affinity model. Estimates for nicotinamide are not applicable because its tight binding is partially because of covalent interaction and the base exchange, which none of the MM-GBSA or LIA or MD methods will properly take into account. Maybe I should include the MM-GBSA numbers for nicotinamide anyways, just so I can explain this to the audience.
- same as above, but for SIRT2
- Done, but the Induced-Fit MM-GBSA results did not complete. There's some type of error occurring and the jobs do not finish after 4 days (way too long, and it the jobs hog most of the licenses). I think I understand how to fix it now. Even if I don't, the LIA results are very good.
- sequence alignment with structural alignment showing SIRT1 & SIRT3 showing the highly conserved residues, residues in the c-pocket and in contact with NAD+. This slide should explain some of the important differences which may contribute to the competitive vs. non-competitive inhibition.
- Slide: "SIRT3 Loops and Residues". Oops, I highlighted residues that were NOT highly conserved. This is easy to change to the opposite.
- More detailed methods: what is necessary to predict binding affinity? what methods? what challenges? What other methods? what have others done? Even more technical details in extra slides that we may not have time to present, but will be helpful with the paper or questions from the audience.
- Also added figure for why I don't need to do loop minimization for the flexible loop in SIRT3. ("loop minimization" titled slide)
- poses and analysis of tight binders of SIRT3, and how they compare to published analysis of SIRT2 or Sir2
- During the paper presentation, when I get to my section of the presentation, I will explain why I ran simulations with Sir2 and SIRT2, rather than just doing simulations with SIRT3. It's because there are more crystal structures available for Sir2, and more experimental data for SIRT2. SIRT3 has limited crystal structures and less experimental IC50 of ∆G_bind.
- Progress with preparation:
- currently working on redoing the SIRT3 simulations with the newest and better crystal structure, PDB:4FVT.
- Also adding ∆G_bind estimates from linear interaction approximation (LIA) method that is also available in the Schrodinger suite and that more literature claims as a better method than MM-GBSA.
- Look for more slide updates on Friday.
- Latest slides are here: Dropbox/pmc-at research/Eric/ACS_slides_03.24.2013.pptx
Fri, Mar 22, 2013
- Draft of ACS presentation slides posted to dropbox here:
- Dropbox/pmc-at research/Eric/ACS_slides.pptx
- Narrowed list of hits to three based on quick availability. See list on Emolecules: "Best 3 from Chembridge Inhibitors to SIRT3"
- ChemBridge supplier IDs: 5281087, 4102009, 9147724
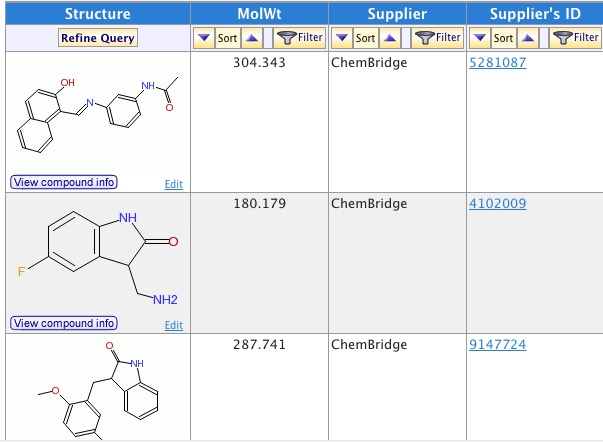
- 5281087 Xiangying already found was available immediately. The other two molecules do not have as good of GlideXP score as 5281087, but are the only ones in the hit set that are possibly in stock from ChemBridge.
- While two of the molecules are a part of a congeneric series, they are not overall a part of a congeneric series. We will need to wait until after the conference to assay a proper congeneric series
- 392 molecules similar to either Salermide, Indole EX-527, or AC-93253 currently being docked. Results will be posted in the afternoon. All molecules are commercially available. Once we have the short list, we can check whether those molecules are in stock.
- List of 392 in Maestro format (can only be opened with Schrodinger software): ligands_similarTo_salermide_EX527_AC93253.maegz
- These molecules were found from the online database of commercially available molecules on www.emolecules.com
- I drew the 3 structures (3 separate queries) , and searched for similar molecules with a similarity score of 0.6 or 0.7
Mon, Mar 18, 2013
- Project 1 update:
- Continued work: as recommended by Schrodinger, multiple input conformations were created with two different force fields (MMFF, OPLS2005) for the set of ligands. Induced Fit docking has been running since Friday, and has an estimated 48 more hours to run for one model with all waters removed of the SIRT2 binding pocket. The job is using the majority of the tokens from our license.
- Finished: Standard GlideXP docking into the SIRT2 crystal structure (PDB:1J8F) and subsequent MM-GBSA rescoring (with minimization of amino acids within 5.0 A from the ligand) did not result in a meaningful correlation between experimental dG_bind and MM-GBSA score (r^2 < 0.1)
- Database queries to www.emolecules.com for the 3 inhibitor molecules Xiangying emailed me (Salermide, Indole EX-527, AC-93253) turned up hundreds of similar molecules. However the similarity index sensitivity needed to be reduced from the default 0.8 to 0.7, which resulted in most of the similar molecules being marginally similar. This means that more than just a single side group is different; i.e., some of the core atoms are different. All of them can be used for docking.
- because Project 1 is using most of the Schrodinger licenses, I think I should pause Project 1 to run a docking job on SIRT3 with these inhibitors.
Thu, Mar 14, 2013
- Just finished conference call with Woody Sherman at Schrodinger.
- Notes of meeting and recommendation on plan:
- Schrodinger.conference.call_2013.03.14.doc
- There were a few questions that I did not get to ask him because he had to cut the meeting to 30 minutes. He said he is available for a follow up next week.
- He also is going to the ACS conference in New Orleans, and is interested in our talk.
While Eric is preparing his notes, I would like to summarize the minutes of the meeting.
1) As far as Lyne paper data is concerned, they used the initial version of Schrodinger software and we use the current version. There could be some difference in the calculation of binding energy between the these two versions. Also, they could have done the various parameter change to improve the R^2 value. This may not be necessary and even a R^2 value of 0.4 is acceptable. Also, he suggested that Lyne, et al could have done some post processing after MM-GBSA docking which could have improved the R^2 value.
2) We have asked how the protein preparation method affects the final results of MM-GBSA simulation - He told that it should not affect much.
3) Is there any stochastic element with Glide simulation so that one should run this multiple times to improve the R^2 value? He said no. but Different computer will handle the simulation differently. For example, Window and Linux based operating system have a different way of rounding number etc. This will affect the simulation results slightly but not very much.
4)He has also suggested to explicitly choose SGB option to dock molecules. We need to learn how to do this Schrodinger software.
Eric will post the detailed notes soon.
Karthik.
RC (3-14): There were several other questions that Eric and I went over on Mon. I hope EK will post on these as well. EK should also follow up tonight with the requested talk/paper planning tasks on the wiki given all the info now in hand.
Wed, Mar 13, 2013
- Meeting with Schrodinger was postponed again by them. They have rescheduled for tomorrow afternoon at 4pm. I will post results of the conversation after the meeting tomorrow.
- Continuing with Project 1 and 2.
Tue, Mar 12, 2013
- Prepared for Schrodinger meeting today by reviewing protocols for the two previous papers in which tried to replicate data, as well as created a list of other questions for them about our larger research goals and which software and how to use it would be best.
- Note: the 5pm conference call was postponed due to a family emergency from the Schrodinger contact.
- Next up:
- finish SIRT2 docking
- based on the email from Schrodinger and my review of the protocols, I can try two more different settings to replicate the data from the Lyne 2006 paper: no post docking minimization in GlideXP, and/or no restraints on the local Prime MM-GBSA minimization.
- Start on project 2 - docking of known SIRT3 ligands for qualitative pose and intermolecular protein-ligand interactions. Talk with Xiangying about which ones she's testing. Dock the set of ligands that are known inhibitors of other sirtuins to find ligands which would be good for Xiangying to run assays on next.
RC (3-13): KM, after the meeting, please post a Q&A listing all questions asked to Schrodinger along with the answers.
Mon, Mar 11, 2013
- Project 1 75% done. Final step to do MM-GBSA calculations.
- Preparing for the Schrodinger meeting. Reviewing the previous protocols and writing questions for tomorrows meeting. Check tomorrow morning for summaries of these.
- Redoing one of the previous paper's calculations (Cardozo) with Macromodel instead of Prime MM-GBSA. Schrodinger said that there is a difference in the two.
Fri, Mar 08, 2013
- Spoke with Karthik for 1 hour about docking and MM-GBSA calculations related to replicating data. How to do it, and helpful hints in using Schrodinger software.
- PDF of interesting new paper Raj pointed out about SIRT1 allosteric activation:
- Working on Project 1 outlined in the 2013.03_strategy.doc Accumulating data from papers, drawing molecules, preparing ligands and protein for docking, etc...
RC (3-8): Eric, I looked over the strategy doc below. Project 1 seems like a good idea and I agree that you should proceed with it. However, we still need to ascertain why we have not been able to reproduce other literature data using MM-GBSA without MD, through discussions with Schrodinger. The discussion points for Schrodinger should be revised before sending out to Schrodinger on Mon. What seems to missing is a description of what freely adjustable parameters were specified in the paper that you were able to reproduce. If you were given a detailed protocol used by the other two papers, what parameters and/or protein/ligand preparation details would have been specified? Regarding treatment of waters, are you planning to redo scoring with these methods prior to speaking to Schrodinger? I would also like to see a plan for how Karthik will be involved in computations for the new strategies.
The backup plan Eric describes in Project 2 is reasonable for the purposes of the presentation, in case we are not able to resolve the issues with Schrodinger. In that case we can start on Project 2 by mid-week next week.
If we can be the first to publish a method of predicting binding affinities of inhibitors to human sirtuin enzymes SIRT3 and SIRT6, it would put us solidly on the sirtuin map.
Thu, Mar 07, 2013
- At library researching more papers related to Sirtuins and computer simulations.
- Downloaded and am reviewing more than a dozen additional papers related to Sirtuins or drug discovery with apo enzymes.
- Most publications that have apo structures do some form of molecular dynamics mixed with other techniques from standard docking to pharmacophore modeling to more MD based ligand binding affinity estimation. What I'm thinking about now is what is the best course of action for us in the next few weeks. List of papers with my analysis of the starting structures, methods and results will be posted.
- More specific strategy outlined here:
- 2013.03_strategy.doc
- This is still a work in progress.
Wed, Mar 06, 2013
- Sent email to Schrodinger. Hopefully they will help us figure out what the problems with replicating the data are.
- continued development of plan for simulations. I realize that Raj wants me to post a very detailed description of the plan for the research to be completed for the ACS meeting. This is critical. I have re-read a number of papers related to this work and am continuing to read others. The issue is that the simple docking with GlideXP then re-scoring with MM-GBSA is a great method that works best under ideal conditions with a co-crystallized structure. Some groups have published results under less ideal conditions, like ours where do don't have a co-crystallized structure of the enzyme with the inhibitor or with the NAD+ cofactor. Most groups use molecular dynamics in these less ideal situations. I need to think and read more before I post this long detailed plan.
Tue, Mar 05, 2013
- Wrote draft of email to Schrodinger requesting help with replicating data.
- Ran a few more iterations of the simulations from the 2006 Lyne paper. Variations involved protein preparation with and without constrained pre-minimization. Alternative strategies did not improve results to agree with the published data.
- Continued to think about strategy for our computations with sirtuins. Our starting crystal structures for SIRT3 do not contain any co-crystallized structures with the inhibitor in the binding pocket. We should follow a strategy like that outlined in the below paper on SIRT2. The authors start with the apo x-ray structure of SIRT2, and use docking then molecular dynamics to elucidate key structural activity relationships in the binding pocket.
- Sakkiah, S., Arooj, M., Kumar, M.R., Eom, S.H., and Lee, K.W. (2013). Identification of inhibitor binding site in human sirtuin 2 using molecular docking and dynamics simulations. PLoS One 8, e51429.
Mon. Mar. 04, 2013
- Revise my simulation part of the paper
- reduce number of figures for simulations
- make finalized high resolution figures for paper
- revise content for simulations
- Good news: my simulation results agree with one of the latest publications with MM-GBSA results.
- Kohlmann, A., Zhu, X., and Dalgarno, D. (2012). Application of MM-GB/SA and WaterMap to SRC Kinase Inhibitor Potency Prediction. ACS Med Chem Lett 3, 94-99.
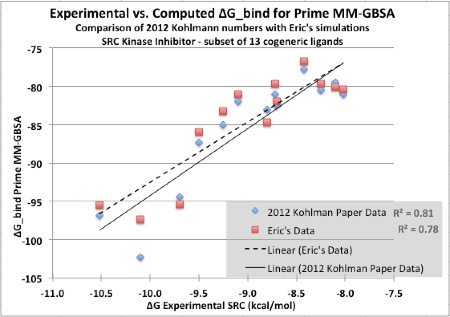 .
.- As the above figure shows, the agreement between the publication and my results are very good. R^2 values for the linear correlation also agree.
- There are a few reasons why this simulation worked so well:
- The paper includes a supplementary section, which explicitly describes the methods and parameters used in the Schrodinger software for their simulations. The other two papers for which I did not get good agreement did not have as detailed a description.
- This paper uses the same major version of the software (Glide 5.7 and Prime 3.0). The other papers used 4+ years earlier versions of the software. Please note that even with the different versions of the software, poor agreement to the reported numbers from the previous 2008 and 2006 publications still should not have been that poor.
- Because of the detailed methods which do NOT mention a restrained pre-minimization of the downloaded PDB structure, I kept the coordinates for the protein atoms fixed from the crystal structure. The previous simulations for the 2006 and 2008 papers did this minimization, which appeared to be a part of the methods in these earlier papers. I am not sure how sensitive MM-GBSA results are to this pre-minimization step.
- This paper explicitly described using MM-GBSA delta_G binding(NS) - that is no strain for the ligand or protein reorganization energy is counted. This is needed to agree with the published results for the 2012 paper. Although using the (NS) values for the other papers still does not create better agreement with published results.
Fri, Mar 01, 2013
- Multiple user connectivity issue to Schrodinger license server resolved.
- Eric and Karthik tested whether we could both simultaneously create an ssh tunnel to the license server from our desktop computers. Previously, Karthik or I had had some problems with the previous slave004 computer where the license server was running. Karthik has a username and login separate from mine on this cluster. We were both able to connect to the server with the ssh tunnel and get permission to run the schrodinger software locally on our desktops. One key is to kill all previous tunnels for the port 27008 (this is the port the license server works on). The previous zombie instances of the ssh tunnel running on the client created a conflict. On the client type the following commands:
ps ax | grep -i ssh# From the output list, note any commands that run the ssh tunnel, similar to step xx below.
# Kill all those processes by finding the process ID number <PID> from the output of ps.
kill -9 <PID>
# restart the ssh tunnelssh -f -N -g -L 27008:75.150.132.105:27008 -L 53000:75.150.132.105:53000 [email protected]
# setup the environmental variables
export SCHROD_LICENSE_FILE=27008@localhost
export SCHRODINGER=/opt/schrodinger/suite2012/ # change the directory to your local installation
# start Maestro
$SCHRODINGER/maestro - This is a bit of a pain. If we involve other students, I can write a shell script to take care of this housekeeping automatically
- Eric and Karthik tested whether we could both simultaneously create an ssh tunnel to the license server from our desktop computers. Previously, Karthik or I had had some problems with the previous slave004 computer where the license server was running. Karthik has a username and login separate from mine on this cluster. We were both able to connect to the server with the ssh tunnel and get permission to run the schrodinger software locally on our desktops. One key is to kill all previous tunnels for the port 27008 (this is the port the license server works on). The previous zombie instances of the ssh tunnel running on the client created a conflict. On the client type the following commands:
- Simulations with the following datasets are currently running on my computer. Due to running each dataset multiple times with various options, each dataset is taking 5 hours to run, not including my analysis. ETA is tomorrow for completion.
- Cdk-2
- Aurora A
- I went through another 10 papers looking for another ideal dataset to test the MM-GBSA software. We have 2 papers so far. I'm still searching for a 3rd. The main problem is that the other papers all use molecular dynamics in conjunction with MM-GBSA. I will report back when I find one with docking + MM-GBSA alone. Hopefully, tomorrow.
Thur, Feb 28, 2013
- Continued additional MM-GBSA simulations from the 2006 J Med Chem 49, 4805-4808 paper. Have done one data set, now doing the other ones. Will report on Friday by 5pm on progress.
- Instructed Karthik to do one of the data sets from this 2006 paper. It will be instructive for him and give me another set of eyes to look at why the numbers are different
- Discussion with Xiangying. She posted some notes of meeting on the "Task List from Lab" site.
- Should we get an MD package such as Amber?
Wed, Feb. 27, 2013
Tasks:
- Other MM-GBSA data sets
my simulations with the Thrombin data sets from the Cardozo paper did exactly replicate their results, we want to confirm that we can replicate MM-GBSA results by testing two other data sets. (a) a different enzyme from the Cardozo paper. (b) a data set from a different paper TBD that shows a correlations between MM-GBSA and experiment - Continue to review literature
- XG reviewed some papers we found a while ago (see her posting from today on Task lists from lab) I should review these papers and post summary.
- Has any one in literature used MM-GBSA and calibrated to experiment to find new high affinity sirtuin inhibitors/activators?
- Not that I found so far.
- update literature. Find new and recent papers not in our list of papers. Review papers
- see below
- put list of docking studies and sirtuin simulations studies in one place. right now the references are scattered throughout the wiki and in the paper references
- in progress.
- Review the J.Med.Chem. v51 (2008) p1203 paper that Xiangying was discussing in meeting on Mon. Feb. 25 on Sir2 docking into C pocket. Specifically pg. 1208 figure
- Note techniques that others are using in publications about docking and MD simulations of sirtuins.
- MD - used in most of newly published studies (< 5 years)
(1) Other MM-GBSA data sets: I did not get results with as high a correlation as the publication.
The best paper that most closely matches what we've been doing with the MM-GBSA re-scoring of the GlideXP docked poses without any molecular dynamics is the following:
- Lyne, P.D., Lamb, M.L., and Saeh, J.C. (2006). Accurate Prediction of the Relative Potencies of Members of a Series of Kinase Inhibitors Using Molecular Docking and MM-GBSA Scoring. J Med Chem 49, 4805-4808.
Here is the info about the ligand set and the MM-GBSA ∆G binding from the publication:
Below are two charts: left is the one I created trying to replicate the one on the right from the paper following their methods using the same Schrodinger software (I used a later version of the software). The paper claims a much higher correlation than I got ( r = 0.84 vs. r = 0.57 for theirs vs. mine).
 |
| My Results |
 |
| Paper |
Below shows the superimposed docked ligands (all 13 of the p38 cogeneric series in this set) after minimization with MM-GBSA. The green molecule is the co-crystallized structure from PDB:2BAK (not in the docked set). As shown, all the ligands in the series docked correctly within 1.5 Å RMSD of the co-crystallized structure, and all are very similar to the co-crysallized structure. Thus, the problem is not related to incorrect docking.
(2) Literature Review
- Neugebauer, R.C., Uchiechowska, U., Meier, R., Hruby, H., Valkov, V., Verdin, E., Sippl, W., and Jung, M. (2008). Structure-activity studies on splitomicin derivatives as sirtuin inhibitors and computational prediction of binding mode. J Med Chem 51, 1203-1213.
- Xiangying wanted my commentary on the methods. This paper, along with many of the other papers that use MM-GBSA to estimate binding affinity, use molecular dynamics (MD) in addition to MM-GBSA to estimate binding affinity. I did not use MD. The MD approach runs a trajectory, then takes 20 to 200 snapshots towards the end of this trajectory from which to calculate MM-GBSA energies to get an ensemble average of the complex, ligand and free protein. This method is much more computationally intensive and time consuming than the method that I used. Recall that I justified the use of the simpler method without MD based in part on these the papers: Guimarães, C. R. W.; Cardozo, M. J. Chem. Inf. Model.2008 and Rastelli, G.; Rio, A. D.; Degliesposti, G.; Sgobba, M. Journal of Computational Chemistry2010, 31, 797.
They claimed that MD did not add accuracy. While it is not clear to me if this is true, especially since I am having difficulty reproducing their results, I still advise that we do use this MD method. Why? Because I suspect that a reviewer or someone who knows simulations will ask why we did not do MD.
- Xiangying wanted my commentary on the methods. This paper, along with many of the other papers that use MM-GBSA to estimate binding affinity, use molecular dynamics (MD) in addition to MM-GBSA to estimate binding affinity. I did not use MD. The MD approach runs a trajectory, then takes 20 to 200 snapshots towards the end of this trajectory from which to calculate MM-GBSA energies to get an ensemble average of the complex, ligand and free protein. This method is much more computationally intensive and time consuming than the method that I used. Recall that I justified the use of the simpler method without MD based in part on these the papers: Guimarães, C. R. W.; Cardozo, M. J. Chem. Inf. Model.2008 and Rastelli, G.; Rio, A. D.; Degliesposti, G.; Sgobba, M. Journal of Computational Chemistry2010, 31, 797.
- Other papers that use MD with MM-GBSA
- Hou, T., Wang, J., Li, Y., and Wang, W. (2011). Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. Journal of chemical information and modeling 51, 69-82.
- Hou, T., Wang, J., Li, Y., and Wang, W. (2011). Assessing the performance of the molecular mechanics/Poisson Boltzmann surface area and molecular mechanics/generalized Born surface area methods. II. The accuracy of ranking poses generated from docking. J Comput Chem 32, 866-877.
- Genheden, S., and Ryde, U. (2011). Comparison of the Efficiency of the LIE and MM/GBSA Methods to Calculate Ligand-Binding Energies. J Chem Theory Comput 7, 3768-3778.
- Rastelli, G., Rio, A.D., Degliesposti, G., and Sgobba, M. (2010). Fast and accurate predictions of binding free energies using MM-PBSA and MM-GBSA. Journal of Computational Chemistry 31, 797-810.
- Hayes, J.M., Skamnaki, V.T., Archontis, G., Lamprakis, C., Sarrou, J., Bischler, N., Skaltsounis, A.-L., Zographos, S.E., and Oikonomakos, N.G. (2011). Kinetics, in silico docking, molecular dynamics, and MM-GBSA binding studies on prototype indirubins, KT5720, and staurosporine as phosphorylase kinase ATP-binding site inhibitors: The role of water molecules examined. Proteins: Structure, Function, and Bioinformatics 79, 703-719.
- Sakkiah, S., Arooj, M., Kumar, M.R., Eom, S.H., and Lee, K.W. (2013). Identification of inhibitor binding site in human sirtuin 2 using molecular docking and dynamics simulations. PLoS One 8, e51429.
- This is an interesting new article for SIRT2
Mon. Feb. 25, 2013
Revised Thrombin simulations. Still not replicating exactly the Cardozo 2008 paper (which had R^2 of 0.7 and 0.8 for single and ensemble MM-GBSA, respectively). The difference between the below and the previously reported thrombin simulations (which had an even larger difference between the published results and my simulations) is that the below fixed the protein docking site, while the previous allowed a zone of flexibility of the protein (all atoms) within 5.0 A of the ligand, as done in the Cardozo paper. Keeping the protein atoms fixed improved the results, but the results still did not match the higher correlation reported in the paper.
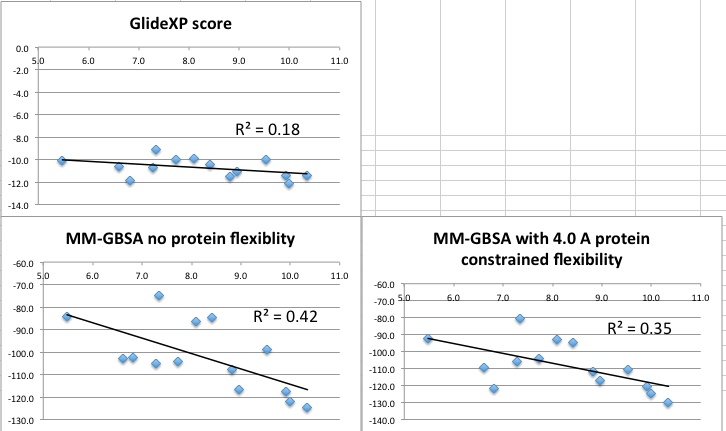
Latest Tasks:
- Due date for tasks: hand calculations and posting of Delta G's of binding for the inhibitors plotted below (Feb 16/17).
Eric, please post the Glide XP and MM-GBSA scores directly in table table above.
EK: done (see below table) - Posting of minutes of EK's discussion on issues pertaining to tasks listed below with RC (Feb 16) RC: Eric, please post this asap (it was due on weekend)
EK: done (see Feb. 16, below) - Posting of results of the thrombin regressions (against computational and experimental literature data) (Feb 18) Eric, this task is due today, with the next three tasks due tomorrow.
EK: In progress. I was very sick Monday with 101 fever while I was finishing the simulations. I think this explains why the results do not match the Cardozo paper's results neither for GlideXP no MM-GBSA. Upon reinspection of the protein model after my fever came down, I realized that the protein preparation wizard incorrectly created a bond in the active site, which caused a propagation of errors. I'm working on fixing this, and redoing the calculations. RC: This is the first I heard about the results not matching Cardozo's results. We should have settled this a couple of weeks ago. Have you done the regression? I assume that this problem was not encountered for the SIRT dockings. But we need to be careful and recheck since a lot of experiments are riding on these results. Also, can you give me an idea how long one such study takes? It would seem it would be possible to complete in one day - hence why the delay? Please comment. There is a lot riding on these results and the 2 week delay is affecting our lab's efforts.EK: these are mostly done. I have to incorporate the globally energy minimized unbound ligand energy into the MM-GBSA numbers. I reported what I have so far. I'm waiting for the - Posting of results of screening of new molecules listed by XG (Feb 19).
RC: Some results on this should be posted prior to our meeting on Wed, with enough lead time for us to look at them, and some commentary requested below. Otherwise the utility of the meeting will be diminished.
EK: First of screening results posted below under Friday, Feb. 22, 2013. More screening simulations in progress. - Posting of commentary of implications of the above results (Feb 19)
- Resolution license server issues (after Feb 19)
EK: New license request sent to Schrodinger on Monday. The new license server is installed and operational on slave003. So far, there have been no conflicts with two users (me and Karthik). However, I still need to run a test with more than 2 simultaneous users before we give access to the Indian students. I will do this next week with Karthik. - Discussion between XG and EK, possibly RC on new experiments needed (Feb 20) EK and XG please provide your preferred time for this meeting
XG: 1:00PM, Feb 20. EK: any time after 2:00pm Feb. 20 is fine. I am not available at 1pm on Feb. 20.
- I would like to reiterate that we have to give an ACS talk on this paper in April and we will not be able to do so properly unless we quickly make substantial progress with these inhibitor studies.
Friday, Feb. 22, 2013
First run of docking to SIRT3 of the database of molecules Xiangying started. The initial list of the molecules is from the document Xiangying created called "Sirtuins inhibitors for NAM binding site_02122013.docx" posted previously on her wiki. These are molecules which have been found in the literature to inhibit activity of a sirtuin or HST enzyme, both human proteins and from other organisms. The screen shown here is when this list was docked with Glide to our model of the SIRT3 binding pocket from PDB:3GLR, which I previously used to dock NAD+ and nicotinamide. Below is a preliminary list of the top Glide scoring molecules (listed by molecule number from Xiangying's original document). This is a preliminary screening of 35 molecules from that list using GlideSP. A more comprehensive screening using GlideXP with more molecules is in progress.
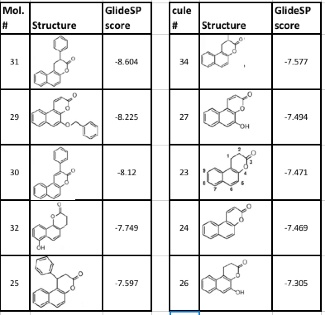
Thursday, Feb. 21, 2013
Thrombin results in table below. Molecule numbers relate to the labeled figure below the table. All columns except the last three are from the Cardozo paper. The last three columns are from my simulations. My simulation numbers are different. Slightly different for GlideXP (within a reasonable error within 2 kcal/mol, relative to each other, given that I used a different version of Glide). My simulation MM-GBSA numbers do not match the Cardozo paper numbers. I need to investigate what happened here. Further below are graphs showing the correlation between my simulations numbers and the reported experimental pKi. These graphs are similar to what was in the Cardozo paper, but the R^2 value is much much lower (mine 0.19 and 0.32, vs. paper was 0.71 and 0.81).
| Molecule # |
pKi |
GlideXP score (kcal/mol) |
MM-GBSA (Ensemble Average) (kcal/mol) |
MM-GBSA (Single Conformer) (kcal/mol) |
XP GScore |
MMGBSA dG Bind (single conformer) |
Relative MM-GBSA (single conformer) |
Relative MM-GBSA (single conformer,corrected with MCMM of unbound ligand) |
| 1 |
7.3 |
-14.0 |
6.1 |
7.2 |
-10.8 |
-51.8 |
70.0 |
239.5 |
| 2 |
7.3 |
-14.3 |
10.0 |
10.3 |
-10.9 |
-107.0 |
14.8 |
126.9 |
| 3 |
8.8 |
-13.9 |
4.3 |
4.9 |
-11.1 |
-109.2 |
12.6 |
79.1 |
| 4 |
6.6 |
-11.8 |
6.0 |
5.9 |
-10.5 |
-100.7 |
21.2 |
73.3 |
| 5 |
9.0 |
-14.0 |
2.1 |
2.3 |
-11.5 |
-106.8 |
15.0 |
52.6 |
| 6 |
6.8 |
-13.3 |
5.8 |
5.6 |
-11.6 |
-114.2 |
7.6 |
74.1 |
| 7 |
5.5 |
-13.2 |
10.9 |
7.9 |
-10.1 |
-90.3 |
31.5 |
107.4 |
| 8 |
9.9 |
-11.2 |
1.9 |
1.9 |
-11.3 |
-118.3 |
3.6 |
21.5 |
| 9 |
10.3 |
-14.1 |
0.0 |
0.0 |
-11.3 |
-121.8 |
0.0 |
13.3 |
| 10 |
8.1 |
-13.1 |
4.8 |
5.6 |
-10.3 |
-103.9 |
18.0 |
5.0 |
| 11 |
8.4 |
-10.3 |
5.2 |
5.9 |
-10.0 |
-105.2 |
16.6 |
0.0 |
| 12 |
7.7 |
-12.7 |
5.3 |
4.2 |
-9.5 |
-98.4 |
23.4 |
124.9 |
| 13 |
10.0 |
-15.7 |
0.7 |
0.9 |
-12.2 |
-119.7 |
2.1 |
32.6 |
| 14 |
9.5 |
-13.0 |
3.3 |
3.6 |
-8.8 |
-84.5 |
37.4 |
19.1 |
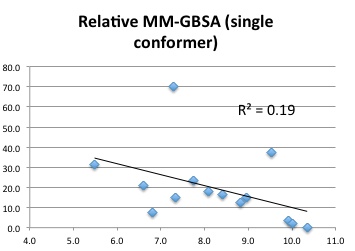

RC: Why have we not rescored these with higher level calculations yet, or otherwise provided some hypothesis as to the cause of the considerable differences in results.
How long does this rescoring take - perhaps a day or less? We cannot operate at a frequency of one update per week on these calculations. How can we proceed with our computational work without resolving this issue asap?
Monday, Feb. 18, 2013
Relationship between K_i and IC_50: linear, with few exceptions, depending on assumptions of kinetic model. The following paper shows detailed equations for the relationship between K_i and IC_50 for Michaelis-Mention kinetics for competitive, non-competitive and uncompetitive inhibitors. It also shows cases for very tightly bound inhibitors. In almost all cases, there is a linear relationship between K_i and IC_50.
- Cer, R.Z., Mudunuri, U., Stephens, R., and Lebeda, F.J. (2009). IC50-to-Ki: a web-based tool for converting IC_50 to K_i values for inhibitors of enzyme activity and ligand binding. Nucleic Acids Res 37, W441–W445. Link to paper.
RC: XG, would you please provide a link to or summary of the method you used to convert IC_50 to K_i.
XG: http://botdb.abcc.ncifcrf.gov/toxin/kiConverter.jsp
RC: Please confirm that that the transformation from IC50 to Ki that you used is linear and provide a short write up on it for the paper if you have not already done so, if it is in fact necessary for obtaining Ki's from the experimental data.
Delta_G of binding for the inhibitors of SIRT3 from Xiangying's experiments.
Assuming Delta_G = RT ln K_i where R=1.99E-3 kcal/(K*mol) and T = 295K, and K_i is converted from uM to molar.
Note that there is no minus sign in the above equation, as binding and inhibition occur in opposite directions. If K_b for binding were used in the equation, there would be a minus sign.
| hSIRT3 inhibition |
Ki(NAD+), uM |
Delta_G (kcal/mol) |
(kcal/mol) |
(kcal/mol) |
||||
| Competitive |
Noncompetitive |
Competitive |
Noncompetitive |
GlideXP |
MM-GBSA |
|||
| Nicotinamide |
11.6 |
-6.66 |
-5.0 |
-31 |
||||
| 1-methylnicotinamide chloride |
2916.3 |
9215 |
-3.42 |
-2.75 |
-4.3 |
-40 |
||
| nicotinic acid N-oxide |
4123.6 |
13030 |
-3.22 |
-2.54 |
4.2 |
-15 |
||
| Iso-nicotinamide |
4367.3 |
13800 |
-3.18 |
-2.51 |
-4.3 |
-33 |
||
| Nicotinic acid |
4588.9 |
14470 |
-3.15 |
-2.48 |
-3.2 |
-22 |
||
| Pyridine N-oxide |
7617.5 |
24070 |
-2.86 |
-2.18 |
-2.2 |
-24 |
||
GlideXP scores are in the correct ball park of -2 to -5 kcal/molWhen comparing these Delta_G numbers to the simulation estimates of Delta_G with the below plots of experiment vs. docking for hSIRT3 inhibitors:
MM-GBSA scores are much lower (-15 to -40 kcal/mol) than absolute binding affinities. However, low numbers like these are reported in many other publications, such as the Cardozo paper.
RC: nicotinic acid N-oxide has a positive glidescore? If not, it is interesting to note that Glide may have been more successful in rank ordering that MM-GBSA. Please comment on this when we meet.
RC: XG, how do the above Ki's for the inhibitors other than NAM compare to others reported in the literature, e.g. for Sir2 or SIRT1, that have received attention as potential drug candidates?
XG: Isonicotinamide was reported as Sir2 activator by relief of NAM inhibition. Nicotinic acid was reported no inhibition effect for Sir2 and SIRT1. I used nicotinic acid as a control. 1-methylnicotinamide chloride, nicotinic acid N-oxide and pyridine N-oxide are from Chief's list, which not published anywhere else in terms of their inhibition effect. Chief wants to test them.
RC: Are there no C pocket inhibitors for SIRT1 that have been proposed as drug candidates? You did not mention any such molecules and their Ki's above.
XG: A quiet few of molecules in the literature (highlighted by red circle below) have good inhibition for hSIRT1. However, it was not clear if they are C pocket inhibitors for hSIRT1 since SIRT1 structure has not been solved to date.
RC: I am trying to understand what is considered a drug candidate for sirtuin inhibition given its Ki. I.e., what binding affinities are we aiming for and is it important to accurately predict the binding affinities of the types of molecules we have assayed so far (or are they orders of magnitude too weak in binding, so we do not need to accurately predict such weak bindingi affinities). Are any of these considered drug candidates? Also, I saw results in the past of inhibitor docking to sirtuins. What pocket did these dock in?
XG: Please comment on this. Can you remind me of what molecules these prior sirtuin docking studies looked at?
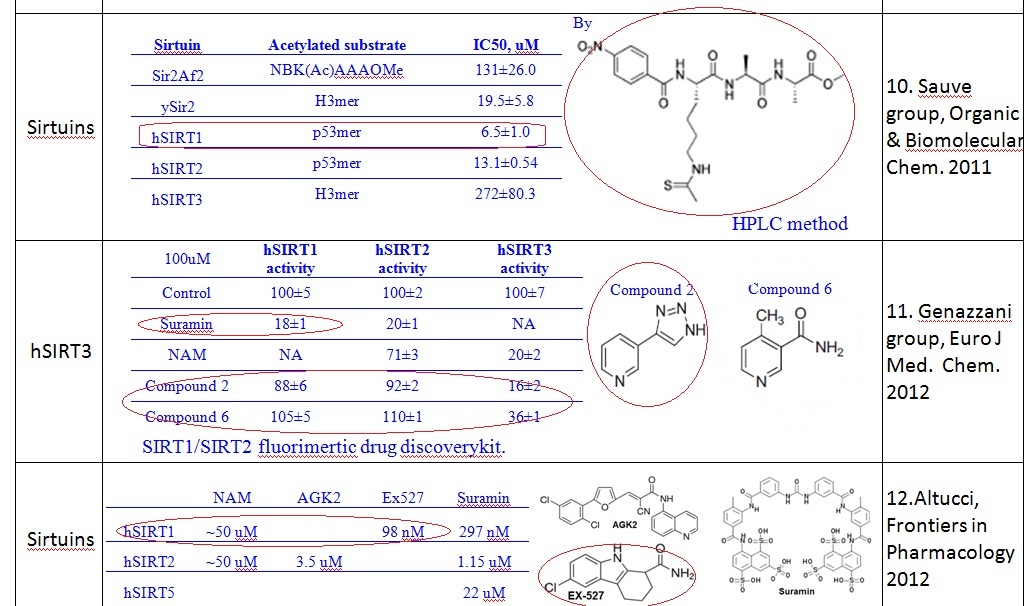
RC: (Sat Feb 16th): I have communicated with Eric on the importance of timely posting of the results requested by me below. We are behind on simulation tasks by a couple of weeks and cannot start on new experiments with inhibitors until these tasks are completed.
Sat. Feb. 16, 2013
Meeting minutes for Talk between RC and EK
- RC: How is IC_50 converted into K_i? Is the relationship between IC_50 and K_i linear?
- EK: (from research after meeting) Yes, in most cases, the relationship is linear. See above from Feb. 18, 2013 notes.
- RC: it is important to calculate and compare delta_G between xiangying's experimental numbers and the simulation numbers.
- EK: see table from Feb. 18, 2013
- RC: for the Thrombin calculations, plot both my simulation numbers vs. the Cardozo simulation numbers. And my simulation numbers vs. the reported experimental numbers.
- RC: we need to finish our simulations and analysis soon for the ACS meeting presentation.
- RC: Eric, you did not post regarding the issues you mentioned with not having a co-crystallized structure, nor regarding the importance of finding higher affinity inhibitors, nor regarding whether we need to be able to accurately predict the binding affinities of the types of inhibitors we have assayed so far, and how the thrombin results will affect our decisions. What is the plan we discussed for XG's next experiments, based on your data? Please be thorough in commentary here.
Tuesday, Feb. 12, 2013
Experiment vs. Docking of hSIRT3 inhibitors (nicotinamide, 1-methyl nicotinamide chloride, nicotinic acid N-oxide, iso-nicotinamide, nicotinic acid, pyridine N-oxide) for both GlideXP and MM-GBSA. The results do not show a clear trend and do not show a better way to estimate the binding affinity through linear regression of docking binding affinity estimates to experimental binding affinity.
RC: Eric, it is important that we post the regression for thrombin inhibitors asap as we have been discussing over the last two weeks. The reason is that we need to assess how well your MM-GBSA scores correlate with the scores reported in the literature before proceeding much further. You could also do a regression of your MM-GBSA scores against the experimentally measured binding affinities for thrombin inhibitors. If it turns out your scores are consistent with those reported in the literature, we know we should look more closely at our lab protocols, whereas if they are not, we should focus on higher levels of sampling, etc. We already have a reason to be skeptical of our experimental Ki's for the new inhibitors - since they did not bind tightly to the C pocket, the Michaelis-Mention approach to determining Ki may not be valid. So, in parallel we should start computationally screening the new inhibitors Guan posted. We really need to accelerate efforts on both these fronts. I believe the above plots should have taken an hour at most to produce. When can you get us these results? Thanks.
Also, as noted below I indicated we should report the experimental Delta G's of binding not just the pIC50's above, so we get an idea as to whether the experimental and computational Delta G's are of the same order of magnitude.
I assume all these tasks will be complete for the Thursday update. I believe that XG has stopped working on inhibitors until we have a next set of tasks from EK, and I would like her to come back to inhibitors next week. We can only do so if there are enough lead compounds identified for lab testing.
Next steps to post by Thursday to redo these graphs with, hopefully, better simulation numbers which show a clearer trend:
- finish the Induced Fit for the six inhibitors.
- run MM-GBSA with higher level of sampling
- also post the thrombin numbers.
Tuesday, Feb. 5, 2013
MM-GBSA literature for re-scoring docked poses. Overall, a handful of papers claim that MM-GBSA re-scoring of docked poses has better correlation to experimental binding affinities. However, the majority of the citations for MM-GBSA and binding affinity relate to higher level simulations where MM-GBSA is used with better sampling techniques such as MD or Monte Carlo. Conclusion: we really need to do the higher level sampling method. The Schordinger software has built in methods for that, such as MCPRO+.
MM-GBSA_literature.doc
Next steps:
- use GlideXP with Induced Fit for the six inhibitors of SIRT3.
- Run MM-GBSA with higher level sampling method, such as MCPRO+ for the 6 ligands.
RC: A little confused here on the status, please help me catch up. Are we ready to do a linear regression of your current MM-GBSA scores against XG's experimentally determined binding affinities? Not sure why we would not want to do this before proceeding with higher level sampling methods. Please let me know asap if you would like to discuss.
EK: Yes, we're ready for the regression. I will post that Tues.
RC: Ok, you can use Delta G = -RT ln K_I (technically, K_I/[C_0] where [C_0] is 1 M). This is - \Delta G_bind since K_I is a dissociation constant so report the negative or use 1/K_I above). This will allow us to compare the actual magnitudes of the experimentally determined \Delta G's to those from computations. (Alternatively, you can use pK_I, as in the literature papers, but these would not be directly comparable to computational values).
RC: Also, why did we not run a linear regression of our MM-GBSA computed thrombin binding affinities against the reported thrombin binding affinities? I posted regarding this last week.
EK: Yes, I will post that Tues, as well.
RC: Please provide the Glidescores and MM-GBSA scores for binding in the other pocket, along with the list of which molecules docked in other pockets. It is obviously important that some of these molecules did not dock into the C pocket in the highest ranked poses, since the experimentally obtained binding affinities may correspond to poses in another pocket. Please discuss the results with XG. XG, do have any other molecules handy that might be added to the cogeneric series? XG, once Eric provides the list of those molecules that bound in other pockets, please let us know whether these molecules had lower Ki's than the others, if you have finished assaying them.
XG: I am still doing the experiments (The experiments I did last week did not work well due to some technical problems). I am catching up. From the preliminary results, it looks like some of these 6 small molecules have different inhibition effect of hSIRT1 vs. hSIRT3. I need to double confirm. If it is true, I will need Eric to dock them into Sir2 protein (1YC2 or 2H4F) for comparison. It will be very interesting to address them in our paper. Eric and I had half hour discussion yesterday. So far, within the 6 molecules, experimentally, NAM is the strongest inhibitor of hSIRT1 and hSIRT3, which is agree with Glidescores but MM-GBSA scores. We will talk again on Friday, hopefully I have some handy data to discuss.What about the important info from the other papers on MM-GBSA? It seems these docking runs below should have only taken a day or so.
Monday, Feb. 4, 2013
See the below table for the GlideXP and MM-GBSA scores for poses docked outside of the C pocket. Note that in all cases except one, the MM-GBSA score ranked the C-pocket poses (for a given ligand) higher than any other pocket for that ligand. Also note that the listed scores do not include higher ranked GlideXP scores in non C-pocket poses, which were subsequently ranked lower than those shown in the table when re-scored with MM-GBSA.
Mon. Jan. 28, 2013
The following are results from docking ligands into SIRT3 (PDB: 3GLR). All of the below ligands docked to the C-pocket without constraints. However, some of the ligands had one or two higher ranked poses that were not in the C-pocket. The below table shows the highest ranked scores that docked in the C-pocket. The last two columns show the best ranked MM-GBSA scoring pose that is not in the C pocket. The other pocket for all cases is in between the B and C pockets (just above the C pocket). Those labeled N/A did not have any poses docked into other volumes besides the C pocket. In only one case (Nicotinic Acid) did the pose docked into the other pocket score higher with MM-GBSA than the C pocket pose. Of note, also, is the discrepancy between the GlideXP scores for Nicotinic Acid N-oxide.
| Pocket ----> |
C |
C |
Other |
Other |
| Ligand |
GlidXP |
MM-GBSA |
GlideXP |
MM-GBSA |
| Nicotinamide |
-5.00 |
-30.8 |
N/A |
N/A |
| iso-nicotinamide |
-4.33 |
-32.5 |
N/A |
N/A |
| 1-methylnicotinamide chloride |
-4.28 |
-40.0 |
-2.83 |
-21.1 |
| Pyridine N-Oxide |
-2.20 |
-23.8 |
N/A |
N/A |
| Nicotinic Acid |
-3.24 |
-21.8 |
-2.78 |
-28.3 |
| Nicotinic Acid N-oxide |
+4.15 |
-14.7 |
-3.30 |
-11.9 |
RC: Are these results with MCMM?
Wed., Jan. 23, 2013
Finished:
- Docked the 14 thrombin inhibitors into the protein (PDB: 1ETT) using GlideXP
- Almost done with docking the 5 inhibitors from Xiangying into SIRT3
To Do:
- Rescore the 14 thrombin inhibitors with MM-GBSA, then use the MCMM ensemble method for free ligand
RC: How did your scores compare to those reported in the lit?
- Finish docking the SIRT3 inhibitors and run the MM-GBSA rescoring
- Post summaries and important info from papers on MM-GBSA
Thurs. Jan 17, 2013
Priorities for the next week:
- Repicate the results from the Cardozo paper for the Thrombin cogeneric series, as outlined in the 2013.11.14.doc below.
- Literature review on MM-GBSA cogeneric series
- Dock the 5 to 6 inhibitors that Xiangying is testing experimentally. Dock with GlideXP into the C pocket of SIRT3, then do MM-GBSA calculations, and report these scores. Hopefully there will be a high correlation between the score and the experimentally determined binding affinity, Ki, from Xiangying.
Jan 14, 2013
Outline for next tasks and projects: 2013.01.14.doc
Dr.Raj,
Please find the attached paper and thesis on MM-GBSA. I have downloaded the thesis of Boas and attached the same here.
I will update once again the wiki with my tasks list.
Best,
Karthik
.PHYSICS-BASED DESIGN OF PROTEIN-LIGAND BINDING - Boas thesis.pdf
MM-GB(PB)SA Calculations of Protein Ligand binding free energies.pdfFast and accurate predictions of binding free energies using MM-PBSA and MM-GBSA.pdfAssessing the performance of the MM-PBSA and MM-GBSA methods The accuracy of binding free energy calculations based on molecular dynamics simulations.pdfAccurate Prediction of the Relative Potencies of Members of a Series of Kinase Inhibitors Using Molecular Docking and MM-GBSA Scoring.pdf
Tasks For Karthik
RC: Karthik, please list your tasks based on Sun discussion with Eric. I have attempted to summarize them here:
1) Literature search: a) MM-GBSA of cogenerics - at least 3 new papers. Are binding affinity tables provided?; b) MD binding affinity prediction papers indicating accuracy of binding affinity prediction for ligands of various sizes.
2) You should also be preparing your GBSA presentation and connecting it up with Eric's presentation on sirtuin binding affinity predictions. Your literature search on MM-GBSA scoring of cogenerics will be useful here. He is also planning to include some mention of the accuracy of MD in absolute binding affinity predictions. This again will rely to a certain extent on your literature search. The ppts should cross reference each other so the connection between projects is clear.
3) Add a few slides to RC's existing slide sets on computational enzyme design provided here:
CMU talk.ppt
neb_enzymes.ppt
for the purpose of a slide presentation to Anna on the project "MM-GBSA scoring project for protein design" listed under "Docking Simulations". The enzyme design part of the CMU talk can be augmented if necessary with a few slides from neb_enzymes (e.g., on the model fitness measure, and possibly a couple of more examples of enzymes and sequence distributions), and then 3-4 new slides for the new MM-GBSA project can be added (see project description for details).
KM can work on these protein design slides while refining his GBSA slides. The protein design slides will b esp useful if MD slides not fully developed, or if there are issues with MD accuracy. KM and RC can jointly present these slides at Anna. KM will also present the GBSA slides. EK can present his slides on sirtuin overview and binding affinity calculation. The right order would appear to be: a) sirtuin overview and binding affinity calculations (including any MD), b) GBSA (KM please provide status update), and then c) protein design. RC might be able to provide the introduction to sirtuins based on EK's slides.
Tasks For Eric
- Completed Tasks in Grey
- Tasks to be done or partially done in Black
- Eric please note the policy on the home page regarding tardy submissions.
Wed Dec 26, 2012
Eric will finish (prior to Jan 1) sirtuin presentation and include the current work for finishing this paper in terms of doing MM-GBSA calculations on a small series of cogeneric inhibitors, as well as the next step where we will look at a larger series of cogenerics.
Wed Dec 19, 2012
RC: I went over the Cardozo paper relatively carefully. Here are my thoughts on what we can realistically aim to achieve in this paper's computational section, as well as a breakdown of tasks between myself and Eric, and what should be left for future work (e.g. the MD project). My comments are based on the results and discussion presented in Cardozo.
- Prediction of binding affinities via MM-GBSA requires a training set of experimental data. It is only possible for a cogeneric series binding in the same pocket.
- Most of the work in computational binding affinity prediction (at least in the absence of MD simulations) focuses on improving correlations between predicted binding affinities and experimental Delta G's for cogenerics.
- In Cardozo Fig 9, correlation between MM-GBSA binding affinities and experimental Ki's are presented for cogenerics (the pKi is the -log Ki). Although we do not have the numbers (unless they are in the supporting information - EK check), based on the Ki's we can compute the experimental Delta G_bind's for these molecules (RC's task) and compare their magnitudes to the MM-GBSA scores on the y-axis, to determine the absolute accuracy of prediction in the absence of an experimental training set.
- (EK) there is no supporting mentioned in the paper or on the journal webpage (here) for this paper. Unless we contact the authors, numbers must be estimated from the plots.
- EK can run a few of the same protein-ligand complexes reported in the Cardozo paper. Since no individual MM-GBSA scores or experimental Ki values are reported (they are only reported as graphs with a collection of inhibitors for each protein), I can compare my calculation to the range of values in the graphs in the Cardozo paper. Am I in the same range? Current MM-GBSA calculation in our SIRT3 and Sir2 work show negative values for MM-GBSA binding of NAD+, but the Cardozo values are + for MM-GBSA for all their different protein ligand complexes. ?????
- The inability of MM-GBSA to predict absolute binding affinities originates, in part, in inaccuracies of the MM force field.
- Glidescores may be in more physically reasonable range than MM-GBSA energies, but they are not as good at rank ordering different ligands in terms of their relative binding affinities (since the scores were parametrized for improving pose prediction accuracy for a given ligand)
- We should break down Delta G_bind contributions into intramolecular, E-S interaction, solvation and configurational entropy contributions, in a table. This table should also compare the single conformation and ensemble results (see also below). EK: check if any similar papers other than Cardozo report computed energies in a table. Please post any such papers, or others that look at cogeneric series Delta G_bind prediction, on the wiki. KM can also help you on literature search.
- EK: look at more MM-GBSA papers.
- Cardozo indicates that use of ensemble <Delta E_intra> rather than single conformation E_intra sometimes magnifies (+) contributions to the predicted Delta G_bind. This is since <Delta E_intra> contains contributions from lower energy unbound ligand conformations. This can cause computed free energies to appear more physically unreasonable, but these effects can "cancel" when predicting Delta G_bind's according to correlations.
- It appears some of the Delta G_bind's computed by MM-GBSA in the Cardozo paper are positive, in Fig 9. EK, can you confirm this?
- (EK) Yes, it appears that the MCMM ensemble score is positive - ranging from 0 to +20.
- For sirtuins, we can predict Delta G_bind computationally if we have experimental binding affinities of other cogenerics that bind in the same pocket
- (EK) may be a possibility and would be along the lines of papers that look at a cogeneric series to estimate binding affinity.
- As noted in my latest posting on the Lab Tasks page, we have these binding affinities for inhibitors, not NAD+.
- We may therefore refocus the computational section of our paper on the prediction of Delta G_bind of other NAM/iso-NAM analogs. XG/EK: How many such inhibitors have we obtained Ki's for so far, and can you provide the Ki's here or in the paper so I can look at them? How many more are in our list? XG, please also provide your comments on how long it would take to run say 5 more such Ki determination experiments on new cogeneric inhibitors
- (EK) I don't know everything that XG has done. There may be more experimental data in literature
- We should compare computational/experimental correlations with and without ensemble averages for the NAM series.
- The results of such studies on NAM analogs should also be more accurate since these are smaller molecules
- For a large molecule like NAD+, it is not surprising that MM-GBSA scores are several times larger than the Glidescores, looking at Cardozo Fig 9.
- How do we justify providing the MM-GBSA scores for NAD+, given that we may not be able to use them for binding affinity predictions at this time, and that Glidescore is good at rank ordering relative binding affinities of the same ligand in different binding pockets (e.g., AC vs AB in Sir2)? We should indicate that we so for completeness and for possible future use in binding affinity predictions if we can obtain a cogeneric series.
- We can also use NAD+ as an example of how ensemble corrections are larger for bigger ligands, since the ensemble corrections may be small for the NAM series.
- Once the experimental and computational work (EK; Karthik could also contribute) for the NAM series is done, RC can compute the Delta G's from the experimentally determined Ki's and then predict Delta G's for new inhibitors from the computational/experimental correlation between the log K_i's and the MM-GBSA scores. The accuracy of the predictions can be tested experimentally. With the ability to computationally predict K_i's, we can predict the effects of any concentration of an inhibitor on the reaction rate. This cannot be done via docking alone, and would be a first in the sirtuin literature.
- It is not clear how MD simulations compare in their ability to predict absolute binding affinities without a training set. Cardozo does not comment on this. EK, you may post a paper answering this question; it is important for the follow-up work.KM can also help you on literature search.If MD can do this, we can apply it to prediction of binding affinities of NAD+ to the AB/AC pockets of Sir2/SIRT3 in the follow-up work, and indicate that the focus of this paper was to apply MM-GBSA to inhibitors/activators, where we have experimental Delta G's for some cogenerics.
- what are the error bars for MD predicted binding affinities?
- why? if we use MD to predict binding affinity for NAD+, how accurate will this be? We can't really get experimental values for NAD+ binding affinity to any of the sirtuin proteins without doing some very specific and difficult assays.
- in table of all the GlideXP, MM-GBSA and MM-GBSA MCMM ligand ensemble scores, break down the components of the scores.
- Figure 4 of the Dec. version of our paper has some relative changes in activity of SIRT3 from XG's experiments for 6 inhibitors / activators. Run MM-GBSA & MM-GBSA MCMM ensemble with those same molecules
- For the project with KM and EK: KM and EK can get IC50 or Ki values from experiments published in the literature for Sir2, SIRT3, SIRT1, etc. For each of these, we can create a cogeneric series of inhibitors (or activators) where we have experimental data for some of them. Then run a huge library of compounds through docking and MM-GBSA to get scores. Correlate those scores to the real data, then use that correlation to predict binding affinity from the predicted MM-GBSA score (for those molecules that don't have experimental data).
- Literature:
- MD. Can it provide a good estimate
- MM-GBSA - more examples of results
- EK and RC discussed that KM should do MD simulations instead of students, and students to KM's project of docking the library of inhibitors. Reason, MD is complicated, while docking is easier for students to do. What can the students do?
- literature search for experimental data and inhibitors.
- what type of library and journal access do they have?
- learn docking and MM-GBSA
- each student have a handful of ligands to dock, along with me doing some docking. The sets of ligands overlap between people for double checking numbers.
- literature search for experimental data and inhibitors.
- MD project: if EK gets the MD project protocol worked out, possibly able to give to students.
- EK: update presentation to included details of docking, MM-GBSA, MD
- why doing MD on NAD+ vs MM-GBSA on inhibitors? b/c NAD+ is not part of cogeneric series, has many rotatable bonds, want good estimate of binding affinity so can use with experimental data to predict forward and backward rate constants, vs just a rank ordering of ligands.
Please provide your comments on the above strategy.
Mon Dec 17, 2012
RC: Eric, after finishing the slide presentation, can you please comment on which of the following priority tasks below you can work on without the licenses:
1b) One thing to check is to compare the molecular mechanics energy portion of the GlideScore to the molecular mechanics energy from MM-GBSA. These components should be similar. Although GlideScore will have a heuristic scaling factor for the molecular mechanics energy. -- do you have this data stored?
5) Verify the calcuations from the 2008 Guimarães and Cardozo paper, “MM-GB/SA Rescoring of Docking Poses in Structure-Based Lead Optimization”.
a) They used an earlier version of MacroModel for all their MM-GBSA and MCMM ensemble calculations.
b) Try to replicate their data.
-- you can't run these yet, but can you make a list of which computations you will run?
7)
b) Reword mentions of “ensemble” at the beginning of the methods section. Ensemble refers to multiple different methods in this section, which is confusing.
c) Put in latest computational simulation #’s for MM-GBSA with MCMM ensemble corrections. RC thinks it would be better if we had one document with the latest results.
d) There are many grammatical errors to correct
f) Length issues: what is the length restriction in JMB (journal of molecular biology)
- (EK) According to the JMB Authors Guide: Articles should normally be no longer than 15 printed pageswith no more than 10 figures and four tables. Additional details about methods notes and supplemental material follows.
- How many long is "15 printed pages"? Here is my estimate.
- Assumptions: 15 printed pages means in the same format as the print edition of JMB, and these 15 pages include the abstract, bibliography and figures
- Based on an analysis of PDF files from >2010 of JMB articles, one page has two columns of text, and a full column of text without any figures has 420 - 480 words. Assuming the average of 450 words per column --> 900 words per page (with NO figures). With 15 pages - 4 pages for figures that leave 11*900 = ~10,000 total words, including bibliography and abstract.
- Our total word count for the Dec. 11 version of the paper is 10,700 words (including bibliography and abstract). So we are within the ballpark for the MAX number of words.
- Supplemental data
The acceptance of supplemental material is at the Editor's discretion. Supplemental information must be submitted with the manuscript for review by the editor and referees. Manuscripts must be complete and stand-alone. Supplemental material should complement the printed paper and may include figures and figure legends, tables, supporting data, sequence alignments, primers, derivation of equations, and videos. The availability of supplemental information will be indicated in the printed paper and the supplemental data will be directly linked to the online version of the paper. Reference to the supplemental information may be made at appropriate places in the text.
With the exception of videos, the supplemental information must be submitted electronically in the form of a single PDF file. Very large tabulations of supporting data may be submitted as Microsoft Excel files.
You can continue working on some of the above tomorrow (we can discuss over skype), and in the meantime I will identify some other tasks for the list for me to work on.
Wed, Dec 12, 2012
Notes from Dec. 10, 2012 phone meeting between Raj and Eric:2012.12.10_notes.doc
Dec. 4, 2012
EK:
Computationally predicted binding affinity too high
As I prepared the next draft of the paper, I double checked a nagging worry I had about the relatively large predicted binding affinities of NAD+ into both the AB and AC pockets of SIRT3 and Sir2 which ranged from -84 to -107 kcal/mol. These are values before the MCMM ensemble correction of the unbound ligand. The values are much higher than any experimental value. As a though experiment, what would the Kb (binding equilibrium constant) be if ∆G(binding) = -90 kcal/mol ?
binding: E + I -> EI K(binding), Kb
∆G(binding) = -R*T*ln Kb
=> Kb
exp(∆G/(R*T))R==1.986 x 10^-3 kcal mol^-1 K^-1, T==298K, ∆G==-90kcal/mol> Kb ≅ 1.06 x 10^66 <===
this is an astronomically huge numberThe BindingDB (http://www.bindingdb.org) is a publicly accessible database currently containing > 20,000 experimentally determined binding affinities of protein–ligand complexes, for >110 protein targets. It can give us an idea of the acceptable range of experimentally determined equilibrium constants, which range from 10^-20 < Ki < 10^10 . Since Ki = 1/Kb, a Kb==10^66 is many orders of magnitude out of normal.
However, large computationally determined binding affinity predictions are sometimes reported in the literature. There are other groups which report ∆G(binding) from MM-GBSA below -100 kcal/mol for a minority of molecules in a large test set (see J Comput Chem. 2011 April 15; 32(5): 866–877). These predicted values are off by an order of magnitude from the experimental values. It would also be reasonable to expect that a large and flexible ligand like NAD+ would produce such a large error in the predicted binding affinity. Errors in the ∆G(binding) are magnified in the predicted Kb and Ki values because of the exponential relationship.
MCMM ensemble averages corrected
Given the above large predicted values for ∆G(binding) I have been double checking the calculations. The ensemble corrected MCMM binding affinity energies increase the deltaG binding for NAD+ to the AB pocket of Sir2 from -95.2 kcal/mol (MM-GBSA) to -26.2 kcal/mol, and increase the binding estimate for the AC pocket from -92.6 kcal/mol to -9.8 kcal/mol. This is partly good news because binding affinities are generally in the range below -25 kcal/mol for most ligands, thus the > 90 kcal/mol estimated by MM-GBSA would draw questions from reviewers. Note that Glide docking scores for both the Sir2 NAD+ AB or AC docking was about -11.0 kcal/mol - within the expected range of -25 to 0 kcal/mol that is normal. So it appears that the ensemble average and the associated more comprehensive exploration of the conformational space of the free NAD+ ligand (instead of simply locally minimizing from the bound protein-ligand complex) is extremely important for a large, highly flexible ligand like NAD+. Entropy contributions (estimated at 1.6 kcal/mol for the unbound ligand ensemble only) is small and well within the margin of error for these estimates. The entropy is not included in the above figures.
The numbers support the noncompetitive mechanism for Sir2, in that NAD+ is predicted to preferentially bind to the AB pocket instead competing with the nicotinamide inhibitor for the C pocket. However, the large difference between the AB vs AC NAD+ binding estimate with the MCMM ensemble method (-26.2 kcal/mol, -9.8 kcal/mol, respectively) may cause some concern.
The numbers for SIRT3 don't look as good after the MCMM ensemble correction:
AB AC
MMGBSA_dG_Bind -84.4 -107.9 kcal/mol <---- MM-GBSA predicted binding affinity before ensemble corrections
Ligand_Energy -47.1 -28.4 kcal/mol <--- uncorrected single conformer unbound ligand energy
<E unbound ligand> -167.6 -167.6 kcal/mol <--- ensemble MCMM average unbound ligand energy
======================================================================
MCMM +36.0 +31.3 kcal/molnote:
MCMM corrected energy = (MMGBSA_dG_Bind) + (Ligand Energy) - <E unbound ligand>
I have to get to the bottom of these positive numbers before I proceed, as we cannot report a positive binding affinity. I suspect that there are some important differences in the two different programs (PRIME for MM-GBSA and MacroModel for MCMM ensemble) which are causing this problem. The 2008 paper that reported good results with the MCMM unbound ligand ensemble energy did all calculations in MacroModel. Note that the above reported results are different from what I reported on the wiki on Sept. 20 because of unlabeled values in output files, where one program reports energies in kcal/mol and the other in kJ/mol. I will report back tomorrow about this.
Methods section edited
In the mean time, the paper is revised through the methods section. (attached) Don't read the results section until I resolve the above issue.
Outline of JMB_120312.doc
Edits through the methods section include:
1) elimination of some sentences and shortening.
2) a short ustification of customized induced fit protocol – better sampling
3) description of the ensemble MCMM method for the unbound ligand
4) brief description that PLOP does not do extensive backbone sampling for induced fit
5) reordering of paragraphs to more clearly describe the three different docking protocols.
6) correction about Epik and PROPKA and partial charges. No semi-empirical or QM method was used to determine ligand partial charges. Partial charges are based on the OPLS force field and the Epik determined protonation states.
I'm waiting to add the rest of the changes until I resolve the ensemble MCMM.
RC: Eric, your tasks we discussed regarding starred item insertion into paper and overall revisions of old material should be submitted by Thursday deadline. Thanks.
RC: Eric and XG, I have read your meeting notes and Eric's latest progress notes. Are we planning to include iso-NAM in this paper or the next paper? It seems a lot of your discussion
focused on iso-NAM. Also, Eric, I will be waiting on your final notes on the use of MM-GBSA for pose generation as well as scoring, vs Glidescore for pose generation and then MM-GBSA scoring.
For mechanistic applications like ours, where we are not screening, are we still able to provide a justification (besides computational efficiency) for using Glidescore?
EK: yes, we're planning on including iso-NAM in this paper. GlideScore rather than MM-GBSA is probably a better metric for generating poses. I'll try to find a reference for this argument.
Nov. 16, 2012 (Fri)
- Need more time to finish the paper revision and rest of the tasks for analyzing the rest of the simulations. Updates will be posted as I finish each task over the next few days.
Nov. 15, 2012 (Thurs)
- Respond to Karthik and coordinate plan for projects with Indian students
- Address remaining tasks from RC regarding the Sirtuin paper. These tasks are posted on the Docking Simulations page. So far I have only addressed the starred tasks.
- The new tasks in the meeting notes from skype conversation with Xiangying
Nov. 11, 2012 (Mon)
- A number of starred tasks that RC posted last week were addressed here: Notes_2012.11.12.doc
- Also, meeting notes of the Nov. 12 skype call between Xiangying and Eric with a task list are in the above document.
Nov. 6, 2012 (Tues)
EK: I do not yet see a list of all tasks we discussed this weekend listed here. This is what Risa was referring to. Please post those asap and finish them by Thursday. Also, there was an ** item missing from your email this weekend. Please address that by Th as well. After Th, meet with Guan to discuss.
Strategy for paper. Both strategies are for 2 papers. I vote for strategy 2.
- Strategy 1:
- Negative: might have to wait for months before Xiangying is ready with 2nd assay to publish the next paper with more computational results.
- Paper 1:
- Experimental: current experimental results from Xiangying, but not including the ongoing continuous assay.
- Computational: only include the docking studies to show modes of binding, but not the binding affinities.
- Possible Journals: Biophysical Journal: not quite as quantitative. Might be very little theory.
- Paper 2: Results for SIRT3 continuous assay (2nd assay). The rest of the computational results, including the binding affinity calculations with MM-GBSA
- Strategy 2: all of current simulation and experimental work in paper 1.
- Paper 1:
- Computational: everything up to now. Docking, MM-GBSA binding affinity
- Experimental: 1st assay only of SIRT3
- Possible Journals: Journal of Molecular Biology, PLoS One
- Paper 2:
- Experimental: 2nd assay of SIRT3 (continuous assay)
- Computational: unknown. Possibly add additional work in MD or other….
- Possible problem: results that include mainly 2nd assay not enough for a full paper.
- Paper 1:
Larger List of Possible Journals.
Rank ordering of Journals. Journals are in a few groups.
- Group 1: general journals. The first 4 are very high ranked journals, and it is unlikely we will be able to publish in a journal such as Nature, JACS or PNAS with this current paper. But J.Med.Chem or PLoS One are possible
- Group 2: Biology, with some theory. Journal of Molecular Biology would be a good journal, then Biophysical journal.
- Group 3: Biology, little theory. These journals would not apply to a paper with significant computational sections
- Group 4: Theory/Computational. These journals would be for a mostly computation paper. Listed in order of preference.
| Journal |
Impact Factor (1) |
Impact Factor. (2) |
Description |
Comments |
| Nature |
36.280 |
General |
||
| Science |
31.200 |
General |
||
| J. Am. Chem. Soc. |
9.907 |
General Chemistry |
||
| PNAS |
9.681 |
General |
||
| PLoS One |
4.092 |
General |
Latest trend journal |
|
| J. Med. Chem. |
5.207 |
5.25 |
Theory and Biology |
Paper would need to have results for many drugs / compounds |
| Biotechnology Advances |
7.60 |
8.25 |
Biology, some theory |
|
| J. Mol. Biol. |
4.008 |
Biology, some theory |
||
| Biophysical Journal |
4.22 |
3.65 |
Biology, some theory |
Not quite as quantitative, very little theory |
| Biochemistry |
3.42 |
Biology, some theory |
||
| Cell |
32.40 |
Biology, little theory |
||
| Molecular Cell |
14.18 |
Biology, little theory |
||
| J. Biol. Chem. |
4.77 |
4.77 |
Biology, little theory |
|
| Mol. Biol. of Cell. |
5.98 |
Biology, little theory |
||
| PLoS Comput. Biol. |
5.22 |
Theory/Computational |
||
| J. Comput. Chem. |
4.080 |
4.58 |
Theory/Computational |
|
| J. Chem. Theory Comput. |
5.22 |
Theory/Computational |
||
| J. Phys. Chem. B |
3.70 |
Theory/Computational |
Despite lower ranking, respected journal in physical chemistry |
|
| J. Chem. Inf. Model |
3.882 |
Theory/Computational |
cheminformatics |
|
| J. Comput. Aid. Mol. Des. |
3.39 |
Theory/Computational |
Low ranking |
|
| J. Mol. Graph. Model. |
2.17 |
Theory/Computational |
Low ranking |
(2) impact factor listed on www.researchgate.net if different from (1)
Nov. 5, 2012 (Mon)
Below are some tasks for the Sirtuin paper.
- 1) * *pg 29-34: Clarify specification of which induced fit protocol was used to get these binding affinities. 4/20, 8/10 of docked structures – refer to the same docking protocol?
- Answer: NO, they do not refer to the same protocol. As described in the methods section:
Since SIRT3 had no publically available cocrystallized structures with NAD+ in the AB or AC pockets, the induced fit protocol was used to dock NAD+ into the AC pocket [figure 15, 8/10 docked]. Neither traditional docking nor the induced fit protocol were sufficient to dock NAD+ into the AB pocket, thus a customized induced fit method was used to dock inhibitors/activators and NAD+ into the AC pocket, as described below [figure 14, 4/20 docked].
Thus, the highlighted phrases were added to the captions of figures 14 and 15: - Figure 14:4 out of the top 20 (based on the emodel glide score; colored white) docked the NAD+ into the AB pocket of SIRT3 using the customized induced fit protocol described in the methods section. Similar to the other figure with AC pocket docking from this same simulation, NAD+ in the AC pocket from the co-crystallized structure of 1YC2:B is red. The 2 structures from 1YC2 (chains A and D) with NAD+ in the AB pocket are pink. The rank order of these 4 structures was 11, 13, 17, and 18 with RMSD to the superimposed 1YC2:A NAD+ of 2.18, 1.82, 2.17, 2.48 Å, respectively.
- Figure 15: 8 out of top 10 (based on emodel glide score) docked the NAD+ into the AC pocket of SIRT3 using the standard induced fit docking protocol. Green are these 8 molecules. Red is the NAD+ in the AC pocket from the co-crystallized structure of 1YC2:B. Pink are the 2 structures from 1YC2 (chains A and D) with NAD+ in the AB pocket. The amide from the nicotinamide is pointing in both directions.\
- Answer: NO, they do not refer to the same protocol. As described in the methods section:
- 4) * *Do we have the MM-GBSA binding affinities for iso-NAM?
- Yes. I will add these figures soon.
- 6) * *Bottom of pg 10: further description in methods section on why docking scoring functions rather than MM-GBSA are used for sampling poses (be aware of length).
- You bring up an interesting issue here: why didn’t I use MM-GBSA as the docking scoring function? 3 reasons:
- 1) the other papers used the Glide score first to find the poses, then rescored those poses using MM-GBSA.
- 2) Using MM-GBSA as the scoring function requires possibly time consuming hacking of Glide
- 3) The glide score has been optimized to find correct poses
- Below is the section you referred to. I added the highlighted section.
The scoring functions in Glide and other docking programs are optimized to minimize the RMSD difference between predicted and x-ray determined structures for a large database of co-crystallized protein-ligand structures [FIND REFERENCE FOR THIS], rather than optimized to predict binding affinity. Thus using the standard scoring functions to predict poses, then subsequently re-scoring those poses with MM-GBSA offers better correlation to actual binding affinities as long as the Glide predicted poses are close to the true binding mode.

October 22, 2012 (Mon)
- Continue manual sorting of data. Same tasks as Oct. 18.
- I'm asking for an extension of the Maestro only license, as sorting through the extensive data is too confusing without my integrated notes and the tables from the Maestro interface. I've spent the last few days looking at the files, but I think that I'm wasting time trying to parse binary data files and formats for Maestro. It will be easier to get another copy of Maestro and work from there. I will update you when I have another copy. I'm sure I can get another copy, as I am not asking for any functionality in simulation or calculation, just viewing.
October 18, 2012 (Thurs)
- Sort through the dozens of simulations.
- Finish documenting procedures.
- Verify reasons for discarding inconsistent data.
- Report results and figures in paper.
- With more than 2 GB of data from the last two weeks, it is much more time consuming to sort through the data without the help of Maestro, which I can no longer use. The data is there and all the simulations are done. For example, I have 7 variations of the simulations with the MM-GBSA calculated energies for the PLOP loop optimized Sir2. Each of these simulations has between 4 to 128 output NAD+ calculated energies for the various docked structures. With my notes that were embedded in Maestro tables about each simulation inaccessible, I am reconstructing what I did for each simulation by looking at the raw output files and viewing in VMD. I need more time to sort through it without the native program that organized the data into easy to access tables and interactive viewing. I'll report on Monday.
Mon., Oct. 15, 2012
Eric - The list below looks good. After the reporting of simulation results is completed I would to arrange for a discussion between you I and Karthik regarding next
steps on the follow-up simulation paper for this project. We have received a number of resumes of Indian students to partake in the NSF-funded project on sirtuin modeling and are interviewing them this week.
That grant has funding for license renewal.
EK: sounds great. I am available from 10am-6pm Mon-Fri for a follow up discussion. I want to incorporate the results into the paper and sift through the results before we have this discussion.
- All simulations for this paper must be finished by Tuesday due to the possible expiration of the trial license
- Besides the following, are there any other calculations, simulations I need to do?
- on going re-calculation of the MM-GBSA energies from the PLOP minimized Sir2 structures
- recalculations done. the energies appear to be fixed and consistent with our hypothesis for Sir2. I ran a lot of simulations in the rush before the license expiration, and am carefully going through the mass of data, which I will report by Thurs. There are multiple sets of results from the different simulations. In particular, some of the simulations have been discarded. I want to review the simulation setup to make sure I'm not cherry picking the positive results. As of the last 4 discarded simulations I've reviewed so far, there were legitimate reasons to discard the data, such as incorrect starting structures, improper force field setup, etc.
- more loop predictions for Sir2 and SIRT3 that lead to a more complete picture / explanation in the paper
- similarly, I ran dozens of simulations with the loops for Sir2 and SIRT3. The task for thursday is to create figures and a consistent explanation of important results.
- The majority of simulations are completed.
- on going re-calculation of the MM-GBSA energies from the PLOP minimized Sir2 structures
Thu., Oct. 11, 2012
- It took a few days to fix the software license issue. It is now fixed.
Mon., Oct. 8, 2012
- IMPORTANT: my access to Microsoft Word expired three days ago, which made it difficult to update the paper. I get another copy of Word tomorrow afternoon. Also, the license to the Schrodinger software is malfunctioning. It should not expire until Oct. 15, but the software has not worked due to licensing errors for the past few days. I am working on a solution so that I can finish the tasks, which require some of this software.
- Update (Oct. 11, 2012) Software access issue to Schrodinger software fixed after a few hours of erasing and reinstalling the software and some online help. An important major update (revision 2) that I installed a few days ago seemed to have caused the problem. Everything is working fine now.
- Important: the trial licence I have expires on Monday, Oct. 15, 2012. While we will have the test simulations and all of the below tasks done, you can decide which version of the software you'd like to purchase.
- Microsoft Word: because of incompatibility issues in editing the paper, I was forced to use Microsoft Word. I had been using a trial version for the past 60 days, which expired a few days ago. I have purchased an installed a full version.
- Update paper as per outline below from Thurs.
- Reduce size of computational methods section by writing more concisely.
Thurs., Oct 4 2012
- Outline of additions to the paper given the new experiments (computational and experimental)
- Xiangying's new data from the new assay
- The methods section already includes a description of the continuous enzyme coupled assay (as opposed the the fluorophore assay). While the experiments are ongoing, the only addition would the the data. And, hopefully, the data supports our original hypothesis, in which both assays show the same results for Sir2 and SIRT3. These additions to the paper, if the hypothesis is supported, would mainly be updates to some figures with the additional data.
- Eric's additions
- additions to methods section for the simulations which added the Boltzmann distribution of states for the free ligand in solution. Additions also to the data section with these results. Since this data supports the previous discussion and conclusion, these other section do not need to be altered
- RC: what about the latest on all the loop predictions in Sir2 and SIRT3, besides the free ligand ensemble calculations? There were several threads on these in the task lists below. How and where will they be incorporated into the paper?
- EK: Yes, these also are to be incorporated into the paper as additional supporting information to the main results and discussion.
- Xiangying's new data from the new assay
- Updated abstract for submission to ACS conference
Mon., Oct. 1, 2012
- check and report on the re-calculation of the MM-GBSA energies from the PLOP minimized Sir2 structures
- [[#|incorporate]]entropy into the corrected ensemble average energy for the MM-GB/SA calculations
- other?
Thurs. Sept. 27, 2012
- Get the following articles to figure out if the MCMM method in MacroModel is sampling from a stationary distribution at a fixed temperature
An internal-coordinate Monte Carlo method for searching conformational space George Chang, Wayne C. Guida, and W. Clark Still. Journal of the American Chemical Society 1989 111 begin_of_the_skype_highlighting
 1989 111 FREE end_of_the_skype_highlighting (12), 4379-4386
1989 111 FREE end_of_the_skype_highlighting (12), 4379-4386Conformations of cycloheptadecane. A comparison of methods for conformational searching Martin Saunders, K. N. Houk, Yun Dong Wu, W. Clark Still, Mark Lipton, George Chang, and Wayne C. Guida Journal of the American Chemical Society 1990 112 (4), 1419-1427
RC: These papers seem to suggest the MCMM algorithm is designed to locate global minima and do not indicate that the algorithm is designed to compute ensemble averages. Still, the GBSA application paper that used it may have modified the input parameters to sample more extensively near the global optimum when generating the list of sequences within a specified window of the global optimum. We will make comments about these points in our paper.
Mon, Sept. 24, 2012
- Add entropy term to the MM-GBSA multiple minimum calculation
- I calculated that the free energy entropy penalty (-T∆S) is small at 1.64 kcal/mol. In this calculation, the entropy of the ligand from the protein-ligand complex (the docked ligand) is assumed 0, while the entropy of the unbound state is calculated from the partition function.
- FYI: here is a spreadsheet showing the calculations and simulation details for the ensemble average energy and the entropy penalty
- Why were the MM-GBSA energies for the PLOP minimized Sir2 structures off?
- The PLOP minimized structures need to be redone, as I could not justify those previous numbers. I think the simulation failed to account for the ligand's presence during the loop minimization, thus creating a receptor conformation that did not conform well to ligand docking (at least in the case of AC docking). I'm re-running the loop minimization with some changes to the files to make sure the ligand is accounted for.
- Submit a revision of the paper with
- EK: wait until get feedback from Raj and finish the rest last of the simulations.
Thurs. Sept. 20, 2012
- Perform the Monte Carlo multiple minimum (MCMM) method in MacroModel to create an ensemble average for the unbound ligand state. Incorporate this average into calculating the ∆G_binding, as described in the Guimarães, & Cardozo 2008 paper. Do this for SIRT3 AC and AB docked MM-GBSA, as well as Sir2 AB and AC.
- The Monte Carlo Multiple Minimum (MCMM) method was used, as implemented in the Macromodel conformational search tool in the Schrodinger GUI. OPLS_2005 forcefield, with extended cutoffs (8.0 Van der Waals, 20.0 A electrostatic, 4.0 for H-bonds), no constraints, PRCG gradient minimization, with 100 steps per rotatable bond, and a 5.0 kcal/mol energy window for [[#|saving]] structures
- I did not include the entropy [[#|term in]] the calculation yet, but the above mentioned paper concluded that this term is not critical.
- The inclusion of the ensemble average for the NAD+ in solution decreased the energy difference for AB pocket binding by 6.4 for SIRT3, while changing the energy for AC binding by only 2.3. Thus, it made the energy difference between AB and AC binding greater, where AB binding is -78 and AC is -105.6. This still fits with the competitive experimental results for SIRT3.
- For SIR2, the results showed a similar trend, with AB pocket binding being penalized by the ensemble approach more than the AC pocket binding. In this case, the final numbers ended up making the ensemble adjusted energies closer for the AB and AC binding at 90.2 and 93.4 for AB and AC binding, fitting with experiment for non-competitive inhibition.
- redo the PLOP minimized Sir2 docking that failed last time.
- The docking worked, but the numbers are off. Standard MM-GBSA energy for docking was 192.4 and 86.2 for AB and AC docking. This does not look right and does not conform to what we would expect.
- [[#|Continue]] integrating and shortening the paper into a cohesive narrative, as outlined in the "General Paper Outline.doc" below.
- Look into why the protein energies are so different for Sir2 and SIRT3
- I checked the components of the energy in the output file for Sir2 and SIRT3 and redid the calculation. The numbers come out the same. It appears that it is the result of the different starting crystal structures for the two systems. We are concerned with the relative differences in energies, not the absolute energies calculated by these simulations. These absolute energies for the docked structures are not reliable.
implemented in Macromodel to get the probabilities of the unbound poses (and then use the Shannon expression to compute the entropy). I believe this is worthwhile to do. It is convenient
that the MC algorithm is already in Macromodel and usable through the GUI. However, I have one question before we proceed: does macromodel directly give you the probabilities of the poses,
or does it just report the global minima. I'm asking since it appears to be simulated annealing variant with a particular convergence diagnostic. Does one have to record steps along the
trajectories and parse them with a script. Recall in the case of our MC protein design code, where we were also computing entropies in an analogous fashion, we sampled conformations to get
at probabilities after convergence to stationarity, at which point we sampled trajectories to get at probabilities.
EK: Answer to your questions: yes, it saves steps of the MC trajectories that are within a certain energy window of the lowest states. The final ensemble is then chosen for the lowest 100 states of the MC trajectory. The probabilities are determined from the Boltzmann distribution with respect to the energy. Higher energy states do not need to be included because the occupancy probability is so low that they do not meaningfully contribute to the ensemble average. Note that I may want to look into whether 100 is a sufficient number of low energy states to include.
EK: I looked at a longer simulation with more than 100 lowest energy conformations. Here, a maximum of 247 conformations were within the 5.0 kcal/mol energy window from the lowest energy structure. Structures beyond the 1st 100 lowest energy conformations only contributed 3% to the total based on the Boltzmann distribution.
RC: Ok, this is the same approach we have been using in our entropy calculations with our protein design MC code. The issue I am mentioning is probably not a major concern, but the reason I was asking is that if Macromodel was not sampling from a stationary distribution after convergence, but rather returning conformations from a simulated annealing algorithm, the probabilities calculated based on the Boltzmann distribution using a partition function computed from the sampled trajectories would not be accurate, since the temperature would not be fixed, and the sampling would not be done from the equilibrium distribution. This is why we spent time (perhaps too much) developing convergence diagnostics in our code. In any case, assuming that the sampling used to collect the 100 states is at fixed temperature after annealing, it is important that the fixed temperature is set to the physiological temperature. Again I assume this is true, but it depends on whether Macromodel sampling is set up for ensemble calculations or only for minimizations. For some reason the link below is not accessible to me without a subscription, but I assume all these conditions are satisfied since Macromodel sampling appears to be automated for ensemble calculations.
EK: Your above comment could be a problem. In order to find out, I would need to read the papers about the MCMM method referenced in the MacroModel user manual. Links to these are in the task list above for someone with journal access to forward to me. Based on the abstracts for these two papers, I do not think that this MCMM method is sampling from a stationary state. Even if the distribution of low energy states is biased because they do not come from a stationary distribution, it is still better than relying on a single conformation, as demonstrated in the "MM-GB/SA Rescoring of Docking Poses in Structure-Based Lead Optimization" paper.
However, additional steps were taken to ensure that the stochastic search approached convergence: an extended protocol for torsional sampling was used, as well as energy minimization to a low gradient norm were employed. That is what the paper did, and those are the options I selected in MacroModel.
Are you computing the ensemble average energies above with \sum_i p_i E_i where p_i = \exp(-E_i/kbT)/Z, with Z = \sum_i \exp(E_i/kbT)? Or are you simply taking the sample average? Is this calculation done by Macromodel or did you write a short script for it? What about for S = -kb \sum_i p_i \ln p_i?
EK: Yes, these are the equations that I used, which assume a normal Boltzmann distribution. I copied 100 of the values from the output files to an excel spreadsheet to calculate. I will look to see if MacroModel does this automatically.
One thing we need to think about is whether these calculations belong in the Biochemistry paper and whether given its length and computational content that is typically not published in Biochemistry we should spin off another paper or submit to a more quantitative journal. We may consider having a discussion next Fri (depending on where XG is with her work).
EK: there might be enough material for a spin off paper for the computational stuff. Although, corrections, for example, of the Boltzmann distribution to a true stationary distribution may not significantly affect the final energy numbers to justify a paper (for this portion). We'll see if we recalculate based on a truly correct ensemble average energy.
Thurs. Sept. 13, 2012
- Investigate ligand reorganization energy importance. Need to get the following paper which discusses this issue with respect to MM-GBSA: Cristiano Guimarães, & Mario Cardozo. (2008) MM-GB/SA Rescoring of Docking Poses in Structure-Based Lead Optimization. Journal of Chemical Information and Modeling, 48(5), 958-970. DOI: 10.1021/ci800004w
Here is the pdf: MM-GB SA Rescoring of Docking Poses in Structure-Based Lead Optimization Journal of Chemical Information and Modeling.pdf- authors looked at cogeneric series of inhibitors for a few systems: HIV-RT, CDK2, FactorXa, & Thrombin
- MM-GB/SA scoring was superior to standard docking scoring functions and the correlation to experimentally determined binding affinities was as good as computationally more expensive Free Energy Perturbation and Thermodynamic Integration (at least for Rank-Ordering ligands based on binding affinity)
- MM-GB/SA calculated ∆G_binding with the single ligand unbound conformer is still much better than standard scoring functions.
- The current MM-GB/SA method I used calculates a single unbound ligand state as a simple energy minimization from the bound state rather than an ensemble average of all possible low energy unbound conformations. This could be particularly misleading because the unbound state for the AC docked pose would have a different energy than the unbound state for the AB docked pose. The unbound energy should be the same for both of these structures.
- The entropy term was for ∆G_binding was found to be relatively unimportant for the tested cogeneric series.
- Ensemble averages for the ligand unbound state are important for more flexible ligands. NAD+ is a very flexible ligand. The factor most affected by ensemble vs. single conformation for the unbound state is the ∆E_intramolecular for the ligand.
- Redo AC docking into SIRT3 without any PLOP sidechain/backbone minimization. Get MM-GBSA energy of this complex.
- -120.3: MM-GBSA ∆G binding for NAD+ docked into the AC pockets of 3GLT using GlideXP, and no PRIME minimization or Induced Fit
- This is lower in energy (better) than the calculated MM-GBSA ∆G binding for AC pocket docked NAD+ that started with 3GLT that had been optimized using PRIME to fit NAD+ into the AB pocket. That energy was -107.9
- The better -120.3 value still fits with the narrative given for the competitive inhibition mechanism for SIRT3. As long as the AC calculated ∆G binding is much lower than that of the AB binding, it fits the competitive inhibition model.
- Notes for me on the simulation: project: 3GLT_hyp1a, entries 237 and 238, which used entry 24 as the receptor (3GLT that had been through the basic protein preparation protocol, and had the intermediate sulfur bond to make a separate acetyl-lys peptide from the thio-intermediate). Possible thing to check: was this acetyl-lys peptide also in the PRIME minimized structure?. NAD+ structures were from entries 25 & 26. Options in MM-GBSA: used input ligand partial charges (verify that these charges were epik minimized); sampling method is basic minimization without constraints, but the distance from ligand to sampled flexible residues is "0.0", so no residues should have been sampled.
- -120.3: MM-GBSA ∆G binding for NAD+ docked into the AC pockets of 3GLT using GlideXP, and no PRIME minimization or Induced Fit
- Look into why the protein energies are so different for Sir2 and SIRT3
- PLOP minimization of the full flexible loop failed for Sir2. I have minimized structures for shorter sections at the beginning and end of this flexible loop. I need to now redock and calculate the MM-GBSA energy for these structures.
- something didn't work here, as the docking failed. Loop optimization seems to have excluded the docked NAD+ and put side chains into the volume where NAD+ docks. Must redo with NAD+ in same file as protein, otherwise it does not seem to be included in the simulation when it is a separate entry.
- my notes: project Sir2_loop_minimize entries 4, 6, and 7-11.
- Skype with Xiangying about paper (issues: paper length, organization, figures, cleanup)
- summary of our discussion from Wed. here: General Paper outline.doc
- RC: I will give some thought to the change of title. Other plans look ok for now. I will get engaged with paper writing/revisions after content has finalized.
Mon. Sept. 10, 2012
- Latest paper draft. Minor additions and revisions added. Outline of Biochemistry_091012.doc
- Add discussion about receptor reorganization energy to paper based on Sept. 6 analysis.Is the paper too long? Do we need to [[#|work]] on making it more concise, removing redundant information, or eliminating some parts?
- Do I need to do additional simulations for this?
- EK: Possibly, depending on how important ligand reorganization energy was found to be in literature. Important: this discussion cannot be added to the paper without also discussing ligand reorganization energy. Ligand reorganization energy (difference between ligand in solution and docked into protein) is just as important as the protein reorganization energy. The MM-GBSA calculations I did are based on an ensemble of docked conformation of the ligand, and does not include this reorganization energy. It can be added by considering multiple conformations. However, there was a paper cited in the current draft that said that MM-GBSA works well with a single conformation of the ligand rather than an ensemble of conformations. I justified the single conformation method on this paper. But, if we [[#|start]] talking about protein reorganization energy, we can’t ignore the ligand reorganization energy. In the todo list for Thurs, there is a paper from Amgen that discusses this very issue with MM-GBSA, which should be forwarded to me.
- RC: Ok, Karthik will send it to you. Please summarize the conclusions of that paper for Thurs and then we can decide how to proceed.
- Is the paper too long? Do we need to [[#|work]] on making it more concise, removing redundant information, or eliminating some parts?
- will discuss with Xiangying on Thurs about shortening the paper.
- Add the loop movement picture based RC feedback.
- Added to paper. Figure 18 in above draft.
- Do loop prediction for the large loop. Previously I only did loop refinement for smaller loops (<12 residues). This large loop for Sir2 PRO33-ALA51 requires that the residues in the conserved secondary structure alpha helix be constrained.
- This did not work, or was taking too long on my laptop. I ran loop prediction for smaller sections of this loop. See comment below that starts with “EK: PLOP failed or was taking too long on…”
- See answers to RC's comments in Sept. 6 section below in red.
Thur. Sept. 6, 2012
- Paper revision: Outline of Biochemistry_090712.doc
- Key priority: Get numbers for receptor reorganization energy for Sir2 and SIRT3 induced fit docking
- For Sir2, the energies are as follows:
| Conformation |
MMGBSA |
Protein OPLS_2005 energy |
| AB |
-95.2 |
-9615 kcal/mol |
| AC |
-92.6 |
-11590 kcal/mol |
- The OPLS_2005 energy is that of the entire protein without the NAD+ ligand. Although the large difference between the two numbers appears to show that the AC protein conformation is a much lower energy (which would be a problem), these numbers must be taken in context. These are redocking scores, and the starting crystal structures for the AB and AC are different, and contain different numbers of other ligands (other than NAD+) and different number of water molecules. The entire protein is also in a slightly different conformation. Thus, these two energies are not a fair comparison. I will think about how to get a fair comparison. Suggestions?RC: I don't recall whether any receptor reorganization by PLOP was needed for Sir2. If not, why do we need to compute the total protein energy?
- EK: No receptor reorganization by PLOP was needed for Sir2. I used the crystal structures directly without any reorganization. But, the AB and AC re-docking started with two different crystal structures. One was the crystal structure with NAD+ co-crystallized in the AB pocket, the other with NAD+ co-crystallized in the AC pocket.
- RC: Understood.
- For SIRT3:
| Conformation |
MMGBSA |
Protein OPLS_2005 energy |
| AB |
-84.4 |
1474520 kcal/mol |
| AC |
-107.9 |
same as above |
- Note that the protein only energy is the same for both the AB and AC docked structures because the shown MMGBSA energies come from the same PRIME minimized protein structure for both AB and AC. Glide docked NAD+ into both the AB and AC pockets without constraints from this PRIME minimized structure. NAD+ probably could have been docked to the AC pocket only with the starting crystal structure before PRIME minimization (I did do this, but the quoted MM-GBSA energy is not from this simulation - which maybe it should be?). This starting crystal structure (after protein preparation with basic minimization and H-bond assignment) had an OPLS2005 energy of 1474570 kcal/mol, which is 50 kcal/mol higher than that of the PRIME minimized structure. This higher energy may appear to contradict the hypothesis that AC binding is lower in energy. However, I believe this is not a fair comparison because the starting crystal structure originated from a possibly strained protein conformation of a trapped thio-intermediate, which may have been further relaxed by the PRIME minimization.
- RC: By minimization, do you mean loop and side chain prediction as well?
- EK: By minimization I mean PRIME loop and side chain prediction with the NAD+ in the AB conformation (from the Sir2 AB co-crystallized structure of NAD+) forced into the SIRT3 AB pocket. PRIME (i.e., PLOP) was used to minimize select side chains (with minor backbone movement) that, upon visual inspection, were sterically hindering the superimposed NAD+ in the AB conformation.
- RC: Ok, we should distinguish between minimization and optimization. Minimization typically refers to local gradient-based minimization.
- We should record the binding affinity for AC with minimization but without any loop and side chain prediction as well.
- EK: Good idea. I need to redo this simulation.
- It appears to me that one should not use the loop optimized structure for AC pocket scoring, since that structure was generated by forcing NAD into the AB pocket, so the backbone will not be in the position it really is in when the ligand binds to AC. We should compare this to the minimized, loop and side chain optimized protein in the case of AB binding. By the way, why are the protein energies so much higher then those for Sir2?
- EK: That is a good question, which I was wondering last week as well. Also note that the OPLS_2005 energy is positive here (for SIRT3), while negative for Sir2. The procedure I used to calculate this energy: I ran an Impact Minimization protocol with the OPLS_2005 forcefield, constant dielectric of 1.0, with a long range force cutoff of 10.0 A for “0” steps. This should have simply calculated the starting structure energy, which I looked up in the output file. Thus no minimization was done, just the starting structure energy was calculated with the given force field. The starting structure was that of the final coordinates of the protein system from the final docked structure with the ligand atoms removed. This is everything but the NAD+, and the protein would have the same PLOP minimized structure (if it was PLOP minimized, as in the case of SIRT3, but not for Sir2). I should look into why the energies are so different. It does look suspicious.
- RC: Ok. Please address my other comments in the bullets above as well. Do you agree with them?
- If I understand correctly, your results with protein minimization are consistent with experimental results.
- EK: The MM-GBSA calculated energies above are consistent with experimental results. I cannot say whether the PLOP minimized SIRT3 structures are consistent with experimental results, because there is no crystal structure with NAD+ in either the AB or AC pockets for SIRT3. Thus we cannot claim that this computed structure (side chain movements or the very slight backbone rearrangement) is definitively the correct structure. Only the relative MM-GBSA docked binding affinities are consistent with experiment of a competitive inhibitor situation. Thus, the SIRT3 PLOP minimized structure is plausible.
- Someone please send me a copy of the original MM-GBSA paper. I don't have access to it:
- Huang, N., Kalyanaraman, C., Irwin, J.J., and Jacobson, M.P. (2006). Physics-Based Scoring of Protein−Ligand Complexes: Enrichment of Known Inhibitors in Large-Scale Virtual Screening. J. Chem. Inf. Model. 46, 243–253.
- Supp_Physics-based scoring of protein ligand complexes_2005_Huang.pdfPhysics-based scoring of protein ligand complexes_2005_Huang.pdf
- refine paper - clean up language, prose, grammar
- add figure of backbone loop movement
- do you mean for Sir2 for the two different crystal structures of NAD+ in the AB or AC pockets? This shows some important backbone movement (shown below)
- or for how the template based induced fit protocol for SIRT3 moved some residues out of the B pocket? In this case, there is much more side chain movement, and some smaller changes in the protein backbone..
- other? I meant the latter. Did we already do this?
- EK: Yes, figure 18 added to paper that shows slight backbone movement only for residues 320-324. Other residues were either constrained or did not move.
- RC:Ok.
- Set up the loop predictions. Analogous protocol could be applied to Sir2. --If this is done, please provide details of what residues were loop optimized and what secondary structure they formed. Also, provide a comparison to the effects of minimization alone.
- recall that we have both the AC and AB pocket crystal structures for Sir2. So this would be a kind of cross-docking check with loop refinement to see if one can take the AB pocket crystal structure and transform it into the AC pocket crystal structure with PLOP backbone minimization? In this case, I have not run a loop prediction, because the number or residues in the loop is too large PRO33-ALA51 for PLOP. However, if I can run a long loop prediction with constraints on the helix (see below), this might work. This alpha-helix moves, while the two shorter loop sections on either side of it reconfigure their sidechains and backbone.
- RC: Yes, I meant that we should apply the same protocol we did to SIRT3 to Sir2 to validate the method with known crystal structures. However, since you are indicating there are major differences between the loops in the two cases, it may not be absolutely necessary. Nonetheless, I agree that we can try to do some constrained loop prediction to validate that PLOP does not move the loops into positions that change the docking scores dramatically.
- EK: PLOP failed or was taking too long on my computer to run for the large number of residues 33-51. Yes, there are major difference between the loops between the AB and AC structures. PLOP minimization (with NAD+ fixed from the co-crystallized structure) of shorter loop sections (separately, the beginning of the loop 33-39, then the end of the loop 46-51) when starting in the AB conformation did not dramatically alter the backbone structure. Here, I was not trying to go from AB to AC, or vice versa. I was trying to see how PLOP minimized the co-crystallized structures, hoping that it did not change the structure too much to alter the predicted MM-GBSA binding energy. I still need to calculate the MM-GBSA energies from these PLOP minimized structures.
- RC: Ok.
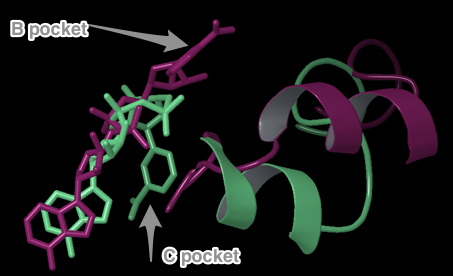
Sir2 from crystal structures. Purple is Sir2 and the NAD+ in the AB conformation. Aqua is Sir2 in the AC pocket. The backbone trace is shown for residues PRO34-ALA51, which is the main loop that moves in the crystal structure.
- RC (8-31): Why are these listed for Thurs rather than Mon? Please list tasks for Mon as well.
- RC (8-31): Had a look over the paper. A few immediate comments:
- a) the SIRT3 figures should explicitly indicate they are for SIRT3 (currently do not indicate which enzyme).I assume this has been fixed. EK: yes.
- b) did we insert the draft commentary we wrote on the Sirtris uncompetitive inhibitor?
- Not yet. This commentary is at the end of the Aug. 23 update here. I will insert it, but should we also try to run a docking simulation with SRT1720?
- RC: No, we shouldn 't run the sim now. EK: OK
- c) Induced Fit (Schrodinger) is referred to throughout, and there is one brief sentence on other approaches to flexible receptor docking. Ideally, we should intersperse some more comments about how other induced fit algorithms behave similarly (e.g., when we say that Induced Fit has limited constraints available or mention other deficiencies).
- Note that I rearranged a few sentences that describe the Schrodinger and other induced fit protocols to emphasize that the Schrodinger protocol is one of many. Also, capitalization of "induced fit" was removed in most instances to emphasize that "induced fit" refers to the general protocol of flexible protein docking, and is not a proprietary name for Schrodingers protocol.
- d) Was the role or lack thereof in loop prediction in Induced Fit mentioned? What was stated about the loop prediction algorithm used by us in our method?
- There is a sentence in the methods section that mentioned loop prediction and the problem with standard induced fit: "For docking NAD+ into the AB pocket of SIRT3, a customized induced fit protocol was used to minimize loops with large steric clashes that the standard induced fit protocol could not accommodate. "
Regarding the loop prediction algorithm: "side chains and backbone residues of the sterically clashing residues were optimized with PLOP."RC: Some more detail on what loop prediction is could be added here.- EK: Added more description: “Side chains and backbone residues of the sterically clashing residues (A:157 to A:160 AND A:320 to A:324, and A:365 to A:367) were optimized with PLOP. This algorithm minimizes only these specified clashing residues around the fixed, superimposed NAD+ in the AB conformation by exhaustively considering sidechain and backbone rotamers based on a rotamer library. Additionally, sidechains are refined for residues within 6.0 Å of the clashing residues, while all other residues remain fixed. A dielectric constant of 1.00 internal and 80.0 external was used. Following this minimization, standard Glide docking is performed without any constraints as previously described.”
- RC: Ok.
- There is a sentence in the methods section that mentioned loop prediction and the problem with standard induced fit: "For docking NAD+ into the AB pocket of SIRT3, a customized induced fit protocol was used to minimize loops with large steric clashes that the standard induced fit protocol could not accommodate. "
- e) I don't think we indicated that in future generations of our method, the docking and loop prediction could be iterated.
- Following sentence added to the "future directions paragraph" at the end of the discussion: "For example, the customized template based induced fit protocol used for docking NAD+ into the AB pocket of SIRT3 could be extended to iteratively incorporate template based loop/side chain prediction with a flexible ligand."
- f) (for XG) Is it correct that the schematics of the modes of inhibition have now been removed and replaced by the equations? Didn't see the schematics in the latest posting.
uploaded the manuscript in dropbox. I proposed that we name our version by date, so that we can add our update by finding the most recent version in either Wiki or dropbox.
Mon. Sept. 3, 2012
- Labor Day
Aug. 30, 2012
- Paper
- Paper revision: Outline of Biochemistry_083112.doc. Note: many changes are highlighted in yellow. There were numerous editing changes below that are not highlighted because noting the multiple changes throughout the document would be confusing.
- Integrate figure numbering
- All figures are now have dynamic numbering and dynamic cross referencing in the text
- add figure of backbone loop movement after optimization
- add figure of NAD+ AC and AB docking for SIRT3? RC: Not sure why this is a question.
- Describe whether/how induced fit was needed to alleviate steric inhibition of the active site Arg in Sir2 (not SIRT3). Added to paper.
- get Xiangyings Endnote Library to integrate with mine.
- We don't need to fully integrate EndNote databases. I figured out that the Word *.doc format includes self contained reference information on all citations, and can dynamically reformat references as they are added, subtracted or moved in the document. While Xiangying and I can work with different citation databases, we both must use Microsoft Word with Endnote for this to work.
- references updated to Biochemistry Journal format (numbered endnotes). This can be changed at any time to another format.
- (EK) Is it OK to cite the Schrodinger Induced fit paper, or should I find some other paper to cite? While I did create my own protocol for docking NAD+ into the SIRT3 AB pocket, Schrodinger's Induced Fit was used for Sir2 and SIRT3 AC.
- RC: You should cite the induced fit paper, but ideally you could cite 1-2 more induced fit papers, since I suspect they would all have the same limitation.
- added citations to a review article on flexible docking using side chain rotamer libraries and one other citation: 'The Induced Fit protocol iteratively uses Glide and a side chain optimization algorithm called PLOP (Jacobson, Friesner et al. 2002) to exhaustively consider possible binding modes and the associated conformational changes within receptor active site in a similar manner as prior algorithms that take advantage of side chain rotamer libraries to add protein receptor flexibility (Schaffer and Verkhivker 1998; Carlson and McCammon 2000). "
- RC (8-21): Do we have the Prime backbone/sidechain reorganization energies handy? Let us add those to the MM-GBSA binding energies since there is an energetic cost to getting the protein reorganized to accept these poses.
- EK: we may not need this, as MM-GBSA includes a ∆E_MM molecular mechanics term. I need to check if this is only the protein-ligand interface, or includes energy terms for all the intramolecular terms within the receptor. If it does, then adding a ligand reorganization energy is redundant. Even so, ligand reorganization energy would still be interesting to report.
- RC: I think you mean protein reorganization energy rather than ligand reorganization energy? Yes, please check on whether MM-GBSA calculates the total energy of the complex or just the interaction energy.
- EK: Concern about the receptor (or ligand...) reorganization energy is important as the ∆G_binding is highly dependent on the cancellation of energy from the receptor-ligand interaction and the energy required to reorganize the receptor and/or ligand. Here we are concerned that the steps I took to change the SIRT3 binding pocket (or for other docking simulations) could create a significant penalty, canceling the energetically more favorable protein-ligand interaction I observed. I looked up a description of MM-GBSA: it does NOT include protein receptor reorganization energy.
Aug. 28, 2012
- Finished adding remaining computational sections to the paper.
- added results from the previous hypothesis in “Sirtuin Kinetics Paper draft 5-16-2012”. Rewrote much of the results section
- full results describing simulations that support experiments showing competitive inhibition of SIRT3 and non-competitive inhibition of Sir2
- removed references to PRIME and other schrodinger products other than Induced Fit and Glide.
- (EK) should I keep the references to the protein prep wizard and associated products?
- re-organized the methods section and added descriptions of the special induced fit protocol used for SIRT3 AB docking. Much of this section was completely reorganized.
- Added paragraph on identifying activators in discussion
- Paper: Outline of Biochemistry paper_082812.doc
- For Risa: same as above paper, but shows changes I made in green. (not to be used for further editing)
- added results from the previous hypothesis in “Sirtuin Kinetics Paper draft 5-16-2012”. Rewrote much of the results section
- Finish full set of nicotinamide and iso-nicotinamide docking. Most have been done, but not for apo-enzyme
Aug. 23, 2012
- catch up on backlog of below tasks. The following tasks from previous lists were moved here as they are completed. RC (8-23): let us move the tasks directly here so Risa can monitor them. Which are to be submitted for Thursday update and which for Mon update?
- start integrating newly written sections with the manuscript draft, and post the updated draft directly here. In progress. Latest version here. Might be better to wait to read until I finish adding all the new sections to the draft on Monday. Outline of biochemistry paper.docx
- RC (8-24): XG and Eric: The paper needs revisions in at least the following two areas to start (in addition to Eric adding all the sections he wrote in response to the tasks on this page): A) we should not refer to Schrodinger suite or Prime protocols by trade name. Instead, -a description of the algorithm- and the name of open source versions of the software should be given (e.g. plop). If this requires reading and citing the original papers from academic groups, that is ok. Glide is only commercial so it is ok to mention it directly. There should not be just one induced fit algorithm cited. There are many available and all share some common deficiencies. More than one should be cited. We did not really use induced fit, but rather our own protocol that consists of loop prediction and docking. This procedure could be iterated to develop a new algorithm and we should propose it briefly.
B) XG needs to insert the equations for competitive, noncompetitive and uncompetitive inhibition into the paper. It is not possible for the reader to understand all the discussion without seeing the equations. There likely should be subsections in the discussion section including ones dedicated to modes of inhibition, and a separate subsection where the computational results are interpreted in the context of these possible modes of inhibition. These have largely been written already, but must be properly positioned in the draft. - Enumerate the roles of the computational (sampling) tools described (in the wiki 8-10 update and in the latest draft) in future studies aimed at answering questions posed in the tasks in “Sirtuin Kinetics Paper draft 5-16-2012” that we decided not to do in this paper. See "Docking Simulations" page, Aug. 23
- Possibly mention the possibility of induced fit docking of the Sirtris uncompetitive inhibitor of SIRT3; mention that since this is an uncompetitive inhibitor, based on the definition of uncompetitive from XG's section, there must be a conformational change that occurs upon NAD+ binding to the AC pocket. In progress. See "Docking Simulations" page, Aug. 23.
Aug. 20, 2012
- Dock NAD+ into the AB pocket from the PRIME modified SIRT3 protein from Thurs., Aug. 16.\ Done, see Aug 21 section of Docking Simulation page on this wiki
- If iso-NAM was not docked, do so. Done, see Aug 21 section of Docking Simulation page on this wiki
- Dock a collection of other molecules into the C pocket in search of other activators / inhibitors ????--
- [[#|step]] one: create a list of > 10 molecules to dock based on previous published inhibitors / activators which Xiangying previously prepared.
- use 3GLT, C pocket as the binding site.
- The following have been in the comments section since last week, and were due latest by yesterday:
- Comment on priorities among remaining tasks listed in the above document (esp iso-NAM docking). XG's draft has a section on iso-NAM. Indicate whether you did iso-NAM docking. Does it bind in the C pocket? See my comments on XG's Mon tasks regarding iso-NAM. See end of Aug 21 section of Docking Simulation page on this wik - See incomplete tasks below - all due Mon.
- All updates should be incorporated into the paper. The revised paper should be posted to the wiki whenever such updates are made with highlighted text indicating where changes were made so Risa can see them.
Due Thurs., Aug. 16, 2012
- Instead of doing MD next, identify flexible loop regions that could be sampled instead of minimized in AB pocket of SIRT3. Set up the loop predictions. Analogous protocol could be applied to Sir2. --If this is done, please provide details of what residues were loop optimized and what secondary structure they formed. Also, provide a comparison to the effects of minimization alone.. This is a prelude to tweaking the induced fit script – based on my previous discussions with you, we would describe how such loop predictions required in the modified algorithm that would be completed later. This includes the ability to combine backbone conformation change energies with binding affinities estimated by MM-GBSA. It may not be the case that simple modifications of the induced fit constraints would help. (completed, see "Docking Simulations" page, section 'Aug. 16')
- Outline the steps in the induced fit protocol (completed, see "Docking Simulations" page, section 'Aug. 16')This was simply copied from another paper and cannot be used by us. We are looking for a write up suitable for our paper - please put it directly in.
- Comment on the challenges inherent in identifying activators via docking methods (e.g., one may need to know the rate of dissociation of iso-NAM,NAM) (completed, see "Docking Simulations" page, section 'Aug. 16')
- change from Endnote to Zotero for all citations in the paper. (completed: my sections are in Zotero format; Xiangying will update her sections with the proper format; see the latest updates to the paper in dropbox).
Due Monday, Aug. 13, 2012
- Increase the number of Induced Fit or Glide output poses to 10,000 or 100,000 in hope that the AB pocket pose would now be included, whereas previously it was rejected because the GlideScore did not meet the more stringent threshold. Did not result in better output. completed 2012.08.13
- Simulated annealing molecular dynamics of SIRT3 with NAD+ constrained in the AB pocket.
- Cycle Computing based FEP molecular dynamics simulation, initial phase - evaluate cycle computing environment.
- Hack Induced Fit script to include additional constraints, evaluation - look at Induced Fit python script to see how difficult it is to understand/modify for this purpose.
- Install MS Office andintegrate drafts with proper footnotes.
- Incorporate comments from “Sirtuin Kinetics Paper draft 5-16-2012” that explain how the simulations we have been doing clarify the possible modes of inhibition (see my comments on XG's Mon tasks regarding modes of inhibition). This will lay the groundwork for cross-referencing of experimental and computational sections in the next draft. Right now there are only 1-2 sentences mentioning how the computational results corroborate the experimental results on noncompetitive inhibition. done. This is is nowhere to be found - there are no references to the extensive document that we spent many weeks preparing and editing; it was a major task that was the priority. XG, please make sure it is done.
- -Split the current discussion section into separate paragraphs describing sampling and energy calculation methods
- Describe whether/how induced fit was needed to alleviate steric inhibition of the active site Arg in Sir2- not exactly sure what RC means here, as part of the current results section focuses on why the induced fit and other increased sampling methods did not work with SIRT3.
- I already commented on this, but it was not addressed. I am talking about Sir2 not SIRT3.
- The remainder of pts listed by RC below under "comments" will be due Thurs.
Due Thursday, Aug. 9, 2012
- See "Discussion of past problems with SIRT3 AB docking and future direction" on the Docking Simulations page.
- identify and briefly describe what types of additional induced fit or dynamics simulations would be required to -properly sample- the side chain and backbone [[#|degrees]] of freedom of SIRT3 in order to quantify the energetics of AB pocket binding, rather than simply indicating that AB poses were not found. This could include a description of current induced fit protocols and their shortcomings. We need to identify to what extent the shortcomings are due to inadequate backbone and sidechain sampling, versus inadequate pose sampling. In Sir2 it was the excluded volumes that enabled successful docking, not backbone or sidechain sampling; this may suggest that an insufficient number of high energy poses were sampled. On the other hand, alleviation of steric clashes in SIRT3 would of course require backbone and sidechain sampling. Is more extensive sampling therefore require in both regards? Can we say anything more specific about the modifications required to the algorithm based on the discussions you and I had? How do excluded volumes focus pose sampling – do they generate more poses in the AB pocket, or is it simply that those AB poses would not be reported otherwise? Dynamics simulations for more accurate estimation of binding affinities should also be mentioned/proposed. (completed 2012.08.09)
Thurs. Aug. 2, 2012 - Tasks for next Monday
- Details on the protocol for docking was completed for today. However, enumerating all the various simulations with a list of exactly which variations of the protocol along with figures of results would be even more helpful to have all results and protocols summarized in one place. See DropBox:PMC-AT Research/Eric/List of docking simulations.doc List is ongoing with updates.
- add references to the and further revisions to the draft. (completed 2012.08.08)
- rewrite Glide and Induced Fit protocols in a form suitable for a paper?? (completed 2012.08.08)
- Add updates to Xiangying's paper draft for the docking simulations. (to be completed 2012.08.08)
- See my additions to the paper on DropBox:Bright Sky/Publications to Be/Sirt.paper.computation.2012.08.07.doc
- Note that the sections in this file can be cut and paste to Xiangying's version. I did not merge the two documents yet because I'm using Open Office and Zotero to manage the references, which will become static and non-updatable in microsoft word. We'll remedy this in a later draft. Should I buy Microsoft Word for ~$90?
Th 8/3
Eric and XG,
Thanks for the Thursday updates.
Eric, I looked at your comparison of the Sir2 and SIRT3 protocols. You did a [[#|good job]] of addressing my questions regarding the steric clashes in SIRT3 that are not present in Sir2. I think that the core docking studies you have done – if clearly explained - should be sufficient for this paper, since there seems to be a simple picture provided for why SIRT3 AB pocket docking is experimentally found to be less favorable than Sir2 AB pocket docking. (However, to be more definitive about this I would need to see the rmsd of NAD+ in the AB pocket of SIRT3 for method 1) – no constraints except single excluded volume.)
Your next steps as indicated above are on mark. I will clarify further here what I would like to see.
The goal for Mon will be to integrate these figures, captions and text into the body of the manuscript.
1) As you indicate beow, we will need to be careful about explaining all steps of the docking protocols, especially those that differ between Sir2 and SIRT3 and why those changes were made. The role of different excluded volumes in Sir2 should be explained. Also, the energy degeneracy of poses differing in the position of NAM should be described – how close in energy are the different NAM conformations, for a given rmsd?
2) We will then compare binding affinities calculated on the basis of the experimental data with the MM-GBSA estimates, and compare different approaches to binding affinity estimation (based on the ppt presentation you gave).
3) [starting Thurs] We will then need to identify and briefly describe what types of additional induced fit or dynamics simulations would be required to -properly sample- the side chain and backbone [[#|degrees]] of freedom of SIRT3 in order to quantify the energetics of AB pocket binding, rather than simply indicating that AB poses were not found. This could include a description of current induced fit protocols and their shortcomings. We need to identify to what extent the shortcomings are due to inadequate backbone and sidechain sampling, versus inadequate pose sampling. In Sir2 it was the excluded volumes that enabled successful docking, not backbone or sidechain sampling; this may suggest that an insufficient number of high energy poses were sampled. On the other hand, alleviation of steric clashes in SIRT3 would of course require backbone and sidechain sampling. Is more extensive sampling therefore require in both regards? Can we say anything more specific about the modifications required to the algorithm based on the discussions you and I had? How do excluded volumes focus pose sampling – do they generate more poses in the AB pocket, or is it simply that those AB poses would not be reported otherwise? Dynamics simulations for more accurate estimation of binding affinities should also be mentioned/proposed.
For the points above, I would like to work with the latest version of the manuscript that XG had been revising – she was aligning the subsections with those of another sirtuin researcher, who I believe was publishing largely experimental enzymology papers in the journal Biochemistry. XG, can you post that latest version with comments on what restructuring has been done. Eric, we will need a complete bibliography to be included with the sections due Monday.
Risa will be looking for the Mon updates. Risa, let me know if you had any trouble accessing the submissions by Eric and XG today.
Raj
Mon. July 30, 2012
Completed [[#|work]] documented in the "Docking Simulations" section of this wiki and in these two documents:
- 2012.08.02_work.docfigures and answers to some of below questions.
- 2012.08.02_docking.protocol.docdetails of the docking protocols.
- PMC-AT wiki setup (completed 2012.07.30)
- make Raj an admin.
- invite Risa.
- send instructions on how to get automatic emails when updates to wiki made.
- Purdue Cluster Move (completed 2012.07.30)
- cleanup and backup my files in my home directory on Purdue cluster in preparation of the cluster move.
- login to new cluster: Public IP: 75.150.132.106
- change default password to a new password.
- Are all my files from old system there? Yes.
- In place MM-GBSA scoring for Sir2 (completed 2012.07.26)
- Additional simulations to get NAD+ docked into the AB pocket of SIRT3 - most failed. (completed 2012.07.31) Still could do additional since SIRT3 docking into AB pocket still fails.
- Annotated figures
- Figures of intermolecular contacts between NAD+ in B vs C pocket for Sir2. (Completed 2012.07.31)
- Similar figure for intermolecular contacts for SIRT3 (only thio-acetyl ADPR intermediate available for SIRT3, which is neither in the A or C pockets) (completed 2012.08.02)
- A previous figure of SIRT3 and Sir2 superpositions was done. Create another more detailed figure showing steric clashes in the B pocket for both sir2 and 3 described and depicted directly in the manuscript . (completed 2012.08.02)
- Write precise protocol that was used for Sir2 and SIRT3 docking (completed 2012.08.02)
- Address Raj's question: was sir2 cross docking was not a "fair test"?
- Comments on Raj's email: "I really don't understand the status with cross docking on Sir2 AB pocket. I'm not sure what we did is a suitable cross-docking test. We need to see the precise protocol" Why was Sir2 cross docking successful and SIRT3 unsuccessful?
- Raj's comment: We need to benchmark any "induced fit" docking methodologies used with SIRT3 on Sir2. Otherwise, it would appear that we have cherry picked the methodology/results.
- Write the status of sir2 AB cross docking. Did it not work? Was the exact same protocol applied to Sir2 and 3? If not why?.
- Add updates to Xiangying's paper draft for the docking simulations. (to be completed 2012.08.01)
- Longer Term Task: Umbrella Sampling or Free Energy Perturbation molecular dynamics simulation.
Questions/Comments
RC: Eric and XG, specific milestones should be posted to the Mon/Thurs section indicating which ones will be submitted by the next deadline. This is how Risa will check progress. Please start by listing the milestones
that will be submitted by Th. This should generally be in the form of some type of attachment that constitutes a deliverable. For Thurs I need to see completed draft versions of bullet points above of "annotated figures" and "write precise protocol" in a form suitable for incorporation (as an attachment). These drafts will be further revised and incorporated into the draft with references and bibliography for next Mon. These are the updates that Risa will check.
RC (8-6): Just an update to indicate that Risa will be checking for Eric's updates first thing Tues am. Thanks.
RC (8-10): The following tasks for Eric are recommended based on his 8-10 update. They should be added directly to the draft. Please ask for clarification where appropriate and post to the Mon task list. Rc will then decide if any should be moved to Thurs. I would not remove points 2,3,4 from Eric's Mon list for now. Points 1,5 should remain. Eric and XG should inspect each other's Mon task lists since they are related.
- Split the current discussion section into separate paragraphs describing sampling and energy calculation methods
- Describe whether/how induced fit was needed to alleviate steric inhibition of the active site Arg in Sir2
- Instead of doing MD next, identify flexible loop regions that could be sampled instead of minimized in AB pocket of SIRT3. Set up the loop predictions. Analogous protocol could be applied to Sir2. This is a prelude to tweaking the induced fit script – based on my previous discussions with you, we would describe how such loop predictions required in the modified algorithm that would be completed later. This includes the ability to combine backbone conformation change energies with binding affinities estimated by MM-GBSA. It may not be the case that simple modifications of the induced fit constraints would help.
- Outline the steps in the induced fit protocol (Thurs)
- Incorporate comments from “Sirtuin Kinetics Paper draft 5-16-2012” that explain how the simulations we have been doing clarify the possible modes of inhibition (see my comments on XG's Mon tasks regarding modes of inhibition). This will lay the groundwork for cross-referencing of experimental and computational sections in the next draft. Right now there are only 1-2 sentences mentioning how the computational results corroborate the experimental results on noncompetitive inhibition.
- Comment on priorities among remaining tasks listed in the above document (esp iso-NAM docking). XG's draft has a section on iso-NAM. Indicate whether you did iso-NAM docking. Does it bind in the C pocket? See my comments on XG's Mon tasks regarding iso-NAM.
- Enumerate the roles of the computational (sampling) tools described (in the wiki 8-10 update and in the latest draft) in future studies aimed at answering questions posed in the tasks in
“Sirtuin Kinetics Paper draft 5-16-2012” that we decided not to do in this paper. Possibly mention the possibility of induced fit docking of the Sirtris uncompetitive inhibitor of SIRT3; mention that since this is an uncompetitive inhibitor, based on the definition of uncompetitive from XG's section, there must be a conformational change that occurs upon NAD+ binding to the AC pocket.
- Comment on the challenges inherent in identifying activators via docking methods (e.g., one may need to know the rate of dissociation of iso-NAM,NAM)
-Risa will be checking Eric's tasks listed for Mon above, even though I have no response from Eric about them. Eric, please note that you will be noted as tardy on Tues morning again if not submitted.